
Top ▲

Gene and Protein Information  |
|||||||
| Species | TM | P Loops | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | 24 | 4 | 2005 | 2q24.3 | SCN2A | sodium voltage-gated channel alpha subunit 2 | 2,48 |
| Mouse | 24 | 4 | 2006 | 2 C1.3 | Scn2a | sodium channel, voltage-gated, type II, alpha | |
| Rat | 24 | 4 | 2005 | 3q21 | Scn2a | sodium voltage-gated channel alpha subunit 2 | 3,36 |
Database Links  |
|
| Alphafold | Q99250 (Hs), P04775 (Rn) |
| ChEMBL Target | CHEMBL4187 (Hs), CHEMBL3399 (Rn) |
| DrugBank Target | Q99250 (Hs) |
| Ensembl Gene | ENSG00000136531 (Hs), ENSMUSG00000075318 (Mm), ENSRNOG00000005018 (Rn) |
| Entrez Gene | 6326 (Hs), 110876 (Mm), 24766 (Rn) |
| Human Protein Atlas | ENSG00000136531 (Hs) |
| KEGG Gene | hsa:6326 (Hs), mmu:110876 (Mm), rno:24766 (Rn) |
| OMIM | 182390 (Hs) |
| Orphanet | ORPHA118500 (Hs) |
| Pharos | Q99250 (Hs) |
| RefSeq Nucleotide | NM_001040142 (Hs), NM_001040143 (Hs), NM_001099298 (Mm), NM_012647 (Rn) |
| RefSeq Protein | NP_001035233 (Hs), NP_001035232 (Hs), NP_001092768 (Mm), NP_036779 (Rn) |
| UniProtKB | Q99250 (Hs), P04775 (Rn) |
| Wikipedia | SCN2A (Hs) |
Selected 3D Structures  |
|||||||||||
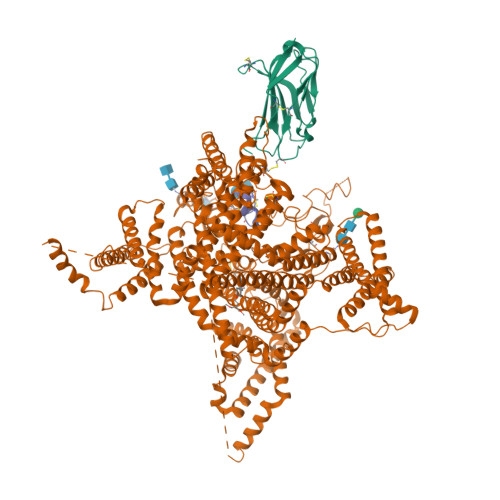
|
|
||||||||||
Associated Proteins  |
||||||||||||||||||||||
|
|
|
||||||||||||||||||||
| Associated Protein Comments | ||||||||||||||||||||||
| β-3 and β-4 subunits also associate with Nav1.2 channels when co-expressed but there are no direct biochemical data on purified sodium channels at present [27,31,50]. | ||||||||||||||||||||||
Functional Characteristics  |
|
| Activation V0.5 = -24 mV. Fast inactivation (τ = 0.8 ms for peak sodium current). | |
Ion Selectivity and Conductance  |
||||||
|
||||||
|
Voltage Dependence  |
||||||||||||||||||||||
|
||||||||||||||||||||||
|
||||||||||||||||||||||
|
||||||||||||||||||||||
| Voltage Dependence Comments | ||||||||||||||||||||||
| The values given for activation and inactivation parameters are for α-subunits expressed alone in mammalian cells. Co-expression of β-subunits shifts the voltage dependence [31], as does the use of intracellular solutions with other major anions (see table). | ||||||||||||||||||||||
Download all structure-activity data for this target as a CSV file 
| Activators | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Activator Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Veratridine and aconitine have been studied on sodium channels in synaptosomes, which are predominantly Nav1.2. Binding affinity for batrachotoxin is greatly enhanced by the presence of co-activators in binding experiments and by depolarising prepulses in voltage-clamp experiments because of high affinity binding to the open/activated state. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gating inhibitors 
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific gating inhibitor tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gating Inhibitor Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Additional scorpion and sea anemone toxins act as inhibitors of fast inactivation of Nav1.2 channels at higher concentrations [20,29,51]. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Channel Blockers | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific channel blocker tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Channel Blocker Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pIC50 values for lidocaine, etidocaine, lamotrigine and phenytoin reflect the Kd values for the resting state of sodium channels are 10 to 100-fold higher than for binding to open and inactivated sodium channels. Many other μ-conotoxins block Nav1.2 channels at higher concentrations [45]. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tissue Distribution 
|
||||||||
|
||||||||
|
||||||||
|
Phenotypes, Alleles and Disease Models 
|
Mouse data from MGI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Clinically-Relevant Mutations and Pathophysiology 
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Biologically Significant Variants 
|
||||||||||||||
|
||||||||||||||
|
||||||||||||||
| Biologically Significant Variant Comments | ||||||||||||||
| The D209/N209 difference in the sequence for these two variants leads to a shift in the voltage dependence of Nav and altered response to epilepsy mutations [36-37,48]. |
1. Abdelsayed M, Sokolov S, Ruben PC. (2013) A thermosensitive mutation alters the effects of lacosamide on slow inactivation in neuronal voltage-gated sodium channels, NaV1.2. Front Pharmacol, 4: 121. [PMID:24065921]
2. Ahmed CM, Ware DH, Lee SC, Patten CD, Ferrer-Montiel AV, Schinder AF, McPherson JD, Wagner-McPherson CB, Wasmuth JJ, Evans GA et al.. (1992) Primary structure, chromosomal localization, and functional expression of a voltage-gated sodium channel from human brain. Proc Natl Acad Sci USA, 89 (17): 8220-4. [PMID:1325650]
3. Auld VJ, Goldin AL, Krafte DS, Marshall J, Dunn JM, Catterall WA, Lester HA, Davidson N, Dunn RJ. (1988) A rat brain Na+ channel alpha subunit with novel gating properties. Neuron, 1 (6): 449-61. [PMID:2856097]
4. Berkovic SF, Heron SE, Giordano L, Marini C, Guerrini R, Kaplan RE, Gambardella A, Steinlein OK, Grinton BE, Dean JT, Bordo L, Hodgson BL, Yamamoto T, Mulley JC, Zara F, Scheffer IE. (2004) Benign familial neonatal-infantile seizures: characterization of a new sodium channelopathy. Ann Neurol, 55 (4): 550-7. [PMID:15048894]
5. Boiko T, Rasband MN, Levinson SR, Caldwell JH, Mandel G, Trimmer JS, Matthews G. (2001) Compact myelin dictates the differential targeting of two sodium channel isoforms in the same axon. Neuron, 30 (1): 91-104. [PMID:11343647]
6. Bricelj VM, Connell L, Konoki K, Macquarrie SP, Scheuer T, Catterall WA, Trainer VL. (2005) Sodium channel mutation leading to saxitoxin resistance in clams increases risk of PSP. Nature, 434 (7034): 763-7. [PMID:15815630]
7. Catterall WA, Morrow CS, Daly JW, Brown GB. (1981) Binding of batrachotoxinin A 20-alpha-benzoate to a receptor site associated with sodium channels in synaptic nerve ending particles. J Biol Chem, 256 (17): 8922-7. [PMID:6114956]
8. Cestèle S, Qu Y, Rogers JC, Rochat H, Scheuer T, Catterall WA. (1998) Voltage sensor-trapping: enhanced activation of sodium channels by beta-scorpion toxin bound to the S3-S4 loop in domain II. Neuron, 21 (4): 919-31. [PMID:9808476]
9. Clare JJ, Tate SN, Nobbs M, Romanos MA. (2000) Voltage-gated sodium channels as therapeutic targets. Drug Discov Today, 5 (11): 506-520. [PMID:11084387]
10. Focken T, Chowdhury S, Zenova A, Grimwood ME, Chabot C, Sheng T, Hemeon I, Decker SM, Wilson M, Bichler P et al.. (2018) Design of Conformationally Constrained Acyl Sulfonamide Isosteres: Identification of N-([1,2,4]Triazolo[4,3- a]pyridin-3-yl)methane-sulfonamides as Potent and Selective hNaV1.7 Inhibitors for the Treatment of Pain. J Med Chem, 61 (11): 4810-4831. [PMID:29737846]
11. Gong B, Rhodes KJ, Bekele-Arcuri Z, Trimmer JS. (1999) Type I and type II Na(+) channel alpha-subunit polypeptides exhibit distinct spatial and temporal patterning, and association with auxiliary subunits in rat brain. J Comp Neurol, 412 (2): 342-52. [PMID:10441760]
12. Hartshorne RP, Catterall WA. (1984) The sodium channel from rat brain. Purification and subunit composition. J Biol Chem, 259 (3): 1667-75. [PMID:6319405]
13. Hartshorne RP, Messner DJ, Coppersmith JC, Catterall WA. (1982) The saxitoxin receptor of the sodium channel from rat brain. Evidence for two nonidentical beta subunits. J Biol Chem, 257 (23): 13888-91. [PMID:6292214]
14. Herlenius E, Heron SE, Grinton BE, Keay D, Scheffer IE, Mulley JC, Berkovic SF. (2007) SCN2A mutations and benign familial neonatal-infantile seizures: the phenotypic spectrum. Epilepsia, 48 (6): 1138-42. [PMID:17386050]
15. Heron SE, Crossland KM, Andermann E, Phillips HA, Hall AJ, Bleasel A, Shevell M, Mercho S, Seni MH, Guiot MC et al.. (2002) Sodium-channel defects in benign familial neonatal-infantile seizures. Lancet, 360 (9336): 851-2. [PMID:12243921]
16. Isom LL, De Jongh KS, Patton DE, Reber BF, Offord J, Charbonneau H, Walsh K, Goldin AL, Catterall WA. (1992) Primary structure and functional expression of the beta 1 subunit of the rat brain sodium channel. Science, 256 (5058): 839-42. [PMID:1375395]
17. Isom LL, Ragsdale DS, De Jongh KS, Westenbroek RE, Reber BF, Scheuer T, Catterall WA. (1995) Structure and function of the beta 2 subunit of brain sodium channels, a transmembrane glycoprotein with a CAM motif. Cell, 83 (3): 433-42. [PMID:8521473]
18. Kahlig KM, Scott L, Hatch RJ, Griffin A, Martinez Botella G, Hughes ZA, Wittmann M. (2022) The novel persistent sodium current inhibitor PRAX-562 has potent anticonvulsant activity with improved protective index relative to standard of care sodium channel blockers. Epilepsia, 63 (3): 697-708. [PMID:35037706]
19. Kaplan MR, Cho MH, Ullian EM, Isom LL, Levinson SR, Barres BA. (2001) Differential control of clustering of the sodium channels Na(v)1.2 and Na(v)1.6 at developing CNS nodes of Ranvier. Neuron, 30 (1): 105-19. [PMID:11343648]
20. Leipold E, Lu S, Gordon D, Hansel A, Heinemann SH. (2004) Combinatorial interaction of scorpion toxins Lqh-2, Lqh-3, and LqhalphaIT with sodium channel receptor sites-3. Mol Pharmacol, 65 (3): 685-91. [PMID:14978247]
21. Liao Y, Deprez L, Maljevic S, Pitsch J, Claes L, Hristova D, Jordanova A, Ala-Mello S, Bellan-Koch A, Blazevic D et al.. (2010) Molecular correlates of age-dependent seizures in an inherited neonatal-infantile epilepsy. Brain, 133 (Pt 5): 1403-14. [PMID:20371507]
22. Linford NJ, Cantrell AR, Qu Y, Scheuer T, Catterall WA. (1998) Interaction of batrachotoxin with the local anesthetic receptor site in transmembrane segment IVS6 of the voltage-gated sodium channel. Proc Natl Acad Sci USA, 95 (23): 13947-52. [PMID:9811906]
23. Liu G, Yarov-Yarovoy V, Nobbs M, Clare JJ, Scheuer T, Catterall WA. (2003) Differential interactions of lamotrigine and related drugs with transmembrane segment IVS6 of voltage-gated sodium channels. Neuropharmacology, 44 (3): 413-22. [PMID:12604088]
24. Mantegazza M, Yu FH, Catterall WA, Scheuer T. (2001) Role of the C-terminal domain in inactivation of brain and cardiac sodium channels. Proc Natl Acad Sci USA, 98 (26): 15348-53. [PMID:11742069]
25. McKerrall SJ, Nguyen T, Lai KW, Bergeron P, Deng L, DiPasquale A, Chang JH, Chen J, Chernov-Rogan T, Hackos DH et al.. (2019) Structure- and Ligand-Based Discovery of Chromane Arylsulfonamide Nav1.7 Inhibitors for the Treatment of Chronic Pain. J Med Chem, 62 (8): 4091-4109. [PMID:30943032]
26. Minassian NA, Gibbs A, Shih AY, Liu Y, Neff RA, Sutton SW, Mirzadegan T, Connor J, Fellows R, Husovsky M et al.. (2013) Analysis of the structural and molecular basis of voltage-sensitive sodium channel inhibition by the spider toxin huwentoxin-IV (μ-TRTX-Hh2a). J Biol Chem, 288 (31): 22707-20. [PMID:23760503]
27. Morgan K, Stevens EB, Shah B, Cox PJ, Dixon AK, Lee K, Pinnock RD, Hughes J, Richardson PJ, Mizuguchi K et al.. (2000) beta 3: an additional auxiliary subunit of the voltage-sensitive sodium channel that modulates channel gating with distinct kinetics. Proc Natl Acad Sci USA, 97 (5): 2308-13. [PMID:10688874]
28. Mulcahy JV, Pajouhesh H, Beckley JT, Delwig A, Du Bois J, Hunter JC. (2019) Challenges and Opportunities for Therapeutics Targeting the Voltage-Gated Sodium Channel Isoform NaV1.7. J Med Chem, 62 (19): 8695-8710. [PMID:31012583]
29. Oliveira JS, Redaelli E, Zaharenko AJ, Cassulini RR, Konno K, Pimenta DC, Freitas JC, Clare JJ, Wanke E. (2004) Binding specificity of sea anemone toxins to Nav 1.1-1.6 sodium channels: unexpected contributions from differences in the IV/S3-S4 outer loop. J Biol Chem, 279 (32): 33323-35. [PMID:15169781]
30. Pan X, Li Z, Huang X, Huang G, Gao S, Shen H, Liu L, Lei J, Yan N. (2019) Molecular basis for pore blockade of human Na+ channel Nav1.2 by the μ-conotoxin KIIIA. Science, 363 (6433): 1309-1313. [PMID:30765605]
31. Qu Y, Curtis R, Lawson D, Gilbride K, Ge P, DiStefano PS, Silos-Santiago I, Catterall WA, Scheuer T. (2001) Differential modulation of sodium channel gating and persistent sodium currents by the beta1, beta2, and beta3 subunits. Mol Cell Neurosci, 18 (5): 570-80. [PMID:11922146]
32. Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. (1994) Molecular determinants of state-dependent block of Na+ channels by local anesthetics. Science, 265 (5179): 1724-8. [PMID:8085162]
33. Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. (1996) Common molecular determinants of local anesthetic, antiarrhythmic, and anticonvulsant block of voltage-gated Na+ channels. Proc Natl Acad Sci USA, 93 (17): 9270-5. [PMID:8799190]
34. Ragsdale DS, Scheuer T, Catterall WA. (1991) Frequency and voltage-dependent inhibition of type IIA Na+ channels, expressed in a mammalian cell line, by local anesthetic, antiarrhythmic, and anticonvulsant drugs. Mol Pharmacol, 40 (5): 756-65. [PMID:1658608]
35. Rogers JC, Qu Y, Tanada TN, Scheuer T, Catterall WA. (1996) Molecular determinants of high affinity binding of alpha-scorpion toxin and sea anemone toxin in the S3-S4 extracellular loop in domain IV of the Na+ channel alpha subunit. J Biol Chem, 271 (27): 15950-62. [PMID:8663157]
36. Sarao R, Gupta SK, Auld VJ, Dunn RJ. (1991) Developmentally regulated alternative RNA splicing of rat brain sodium channel mRNAs. Nucleic Acids Res, 19 (20): 5673-9. [PMID:1658739]
37. Schlachter K, Gruber-Sedlmayr U, Stogmann E, Lausecker M, Hotzy C, Balzar J, Schuh E, Baumgartner C, Mueller JC, Illig T et al.. (2009) A splice site variant in the sodium channel gene SCN1A confers risk of febrile seizures. Neurology, 72 (11): 974-8. [PMID:19289736]
38. Schmalhofer WA, Calhoun J, Burrows R, Bailey T, Kohler MG, Weinglass AB, Kaczorowski GJ, Garcia ML, Koltzenburg M, Priest BT. (2008) ProTx-II, a selective inhibitor of NaV1.7 sodium channels, blocks action potential propagation in nociceptors. Mol Pharmacol, 74 (5): 1476-84. [PMID:18728100]
39. Shi X, Yasumoto S, Nakagawa E, Fukasawa T, Uchiya S, Hirose S. (2009) Missense mutation of the sodium channel gene SCN2A causes Dravet syndrome. Brain Dev, 31 (10): 758-62. [PMID:19783390]
40. Striano P, Bordo L, Lispi ML, Specchio N, Minetti C, Vigevano F, Zara F. (2006) A novel SCN2A mutation in family with benign familial infantile seizures. Epilepsia, 47 (1): 218-20. [PMID:16417554]
41. Stühmer W, Methfessel C, Sakmann B, Noda M, Numa S. (1987) Patch clamp characterization of sodium channels expressed from rat brain cDNA. Eur Biophys J, 14 (3): 131-8. [PMID:2435540]
42. Sutkowski EM, Catterall WA. (1990) Beta 1 subunits of sodium channels. Studies with subunit-specific antibodies. J Biol Chem, 265 (21): 12393-9. [PMID:2165060]
43. Westenbroek RE, Merrick DK, Catterall WA. (1989) Differential subcellular localization of the RI and RII Na+ channel subtypes in central neurons. Neuron, 3 (6): 695-704. [PMID:2561976]
44. Whitaker WR, Clare JJ, Powell AJ, Chen YH, Faull RL, Emson PC. (2000) Distribution of voltage-gated sodium channel alpha-subunit and beta-subunit mRNAs in human hippocampal formation, cortex, and cerebellum. J Comp Neurol, 422 (1): 123-39. [PMID:10842222]
45. Wilson MJ, Yoshikami D, Azam L, Gajewiak J, Olivera BM, Bulaj G, Zhang MM. (2011) μ-Conotoxins that differentially block sodium channels NaV1.1 through 1.8 identify those responsible for action potentials in sciatic nerve. Proc Natl Acad Sci USA, 108 (25): 10302-7. [PMID:21652775]
46. Wollner DA, Messner DJ, Catterall WA. (1987) Beta 2 subunits of sodium channels from vertebrate brain. Studies with subunit-specific antibodies. J Biol Chem, 262 (30): 14709-15. [PMID:2444590]
47. Xie X, Lancaster B, Peakman T, Garthwaite J. (1995) Interaction of the antiepileptic drug lamotrigine with recombinant rat brain type IIA Na+ channels and with native Na+ channels in rat hippocampal neurones. Pflugers Arch, 430 (3): 437-46. [PMID:7491269]
48. Xu R, Thomas EA, Jenkins M, Gazina EV, Chiu C, Heron SE, Mulley JC, Scheffer IE, Berkovic SF, Petrou S. (2007) A childhood epilepsy mutation reveals a role for developmentally regulated splicing of a sodium channel. Mol Cell Neurosci, 35 (2): 292-301. [PMID:17467289]
49. Yamaji N, Little MJ, Nishio H, Billen B, Villegas E, Nishiuchi Y, Tytgat J, Nicholson GM, Corzo G. (2009) Synthesis, solution structure, and phylum selectivity of a spider delta-toxin that slows inactivation of specific voltage-gated sodium channel subtypes. J Biol Chem, 284 (36): 24568-82. [PMID:19592486]
50. Yu FH, Westenbroek RE, Silos-Santiago I, McCormick KA, Lawson D, Ge P, Ferriera H, Lilly J, DiStefano PS, Catterall WA et al.. (2003) Sodium channel beta4, a new disulfide-linked auxiliary subunit with similarity to beta2. J Neurosci, 23 (20): 7577-85. [PMID:12930796]
51. Zaharenko AJ, Schiavon E, Ferreira Jr WA, Lecchi M, de Freitas JC, Richardson M, Wanke E. (2012) Characterization of selectivity and pharmacophores of type 1 sea anemone toxins by screening seven Na(v) sodium channel isoforms. Peptides, 34 (1): 158-67. [PMID:21802465]