|
Synonyms: CI-1003 | suramine
suramin is an approved drug
Comment: Suramin is an antiparasitic drug. It is also used in research as a broad-spectrum P2 receptor antagonist [ 1, 5] and as a ryanodine receptor agonist [ 13]. Evidence from studies in mice [ 7-9], and from an early phase clinical trial suggest antipurinergic therapy (using suramin or other antipurinergic compounds) as a potential lead in the search for pharmacotherapeutic intervention in children with autism spectrum disorder (see Phase 1/2 clinical trial NCT02508259).
|
|
2D Structure
|
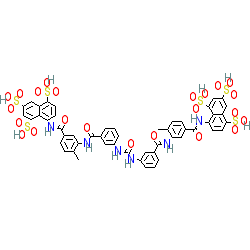
|
Physico-chemical Properties
|
|
|
Hydrogen bond acceptors
|
29
|
|
Hydrogen bond donors
|
12
|
|
Rotatable bonds
|
22
|
|
Topological polar surface area
|
534.03
|
|
Molecular weight
|
1296.05
|
|
XLogP
|
0.91
|
|
No. Lipinski's rules broken
|
3
|
|
SMILES / InChI / InChIKey
|
|
|
Canonical SMILES
|
O=C(Nc1cccc(c1)C(=O)Nc1cc(ccc1C)C(=O)Nc1ccc(c2c1c(cc(c2)S(=O)(=O)O)S(=O)(=O)O)S(=O)(=O)O)Nc1cccc(c1)C(=O)Nc1cc(ccc1C)C(=O)Nc1ccc(c2c1c(cc(c2)S(=O)(=O)O)S(=O)(=O)O)S(=O)(=O)O
|
|
Isomeric SMILES
|
O=C(Nc1cccc(c1)C(=O)Nc1cc(ccc1C)C(=O)Nc1ccc(c2c1c(cc(c2)S(=O)(=O)O)S(=O)(=O)O)S(=O)(=O)O)Nc1cccc(c1)C(=O)Nc1cc(ccc1C)C(=O)Nc1ccc(c2c1c(cc(c2)S(=O)(=O)O)S(=O)(=O)O)S(=O)(=O)O
|
|
InChI
|
InChI=1S/C51H40N6O23S6/c1-25-9-11-29(49(60)54-37-13-15-41(83(69,70)71)35-21-33(81(63,64)65)23-43(45(35)37)85(75,76)77)19-39(25)56-47(58)27-5-3-7-31(17-27)52-51(62)53-32-8-4-6-28(18-32)48(59)57-40-20-30(12-10-26(40)2)50(61)55-38-14-16-42(84(72,73)74)36-22-34(82(66,67)68)24-44(46(36)38)86(78,79)80/h3-24H,1-2H3,(H,54,60)(H,55,61)(H,56,58)(H,57,59)(H2,52,53,62)(H,63,64,65)(H,66,67,68)(H,69,70,71)(H,72,73,74)(H,75,76,77)(H,78,79,80)
|
|
InChI Key
|
FIAFUQMPZJWCLV-UHFFFAOYSA-N
|
|
|

