
Top ▲

GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
Target not currently curated in GtoImmuPdb
Target id: 1256
Nomenclature: protein arginine methyltransferase 5
Abbreviated Name: PRMT5
Gene and Protein Information  |
||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | - | 637 | 14q11.2 | PRMT5 | protein arginine methyltransferase 5 | |
| Mouse | - | 637 | 14 C2 | Prmt5 | protein arginine N-methyltransferase 5 | |
| Rat | - | - | 15p13 | Prmt5 | protein arginine methyltransferase 5 | |
Previous and Unofficial Names  |
| Histone-arginine N-methyltransferase | Jak-binding protein 1 | Jbp1 | HRMT1L5 |
Database Links  |
|
| Alphafold | O14744 (Hs), Q8CIG8 (Mm) |
| ChEMBL Target | CHEMBL1795116 (Hs) |
| Ensembl Gene | ENSG00000100462 (Hs), ENSMUSG00000023110 (Mm), ENSRNOG00000012046 (Rn) |
| Entrez Gene | 10419 (Hs), 27374 (Mm), 364382 (Rn) |
| Human Protein Atlas | ENSG00000100462 (Hs) |
| KEGG Gene | hsa:10419 (Hs), mmu:27374 (Mm), rno:364382 (Rn) |
| OMIM | 604045 (Hs) |
| Pharos | O14744 (Hs) |
| SynPHARM |
81924 (in complex with EPZ015666) 83227 (in complex with GSK591) |
| UniProtKB | O14744 (Hs), Q8CIG8 (Mm) |
| Wikipedia | PRMT5 (Hs) |
Selected 3D Structures  |
|||||||||||||
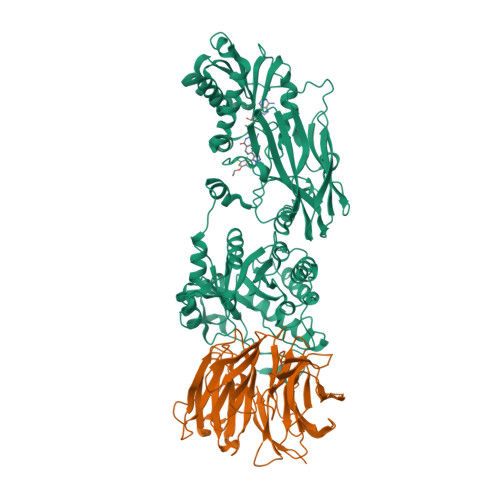
|
|
||||||||||||
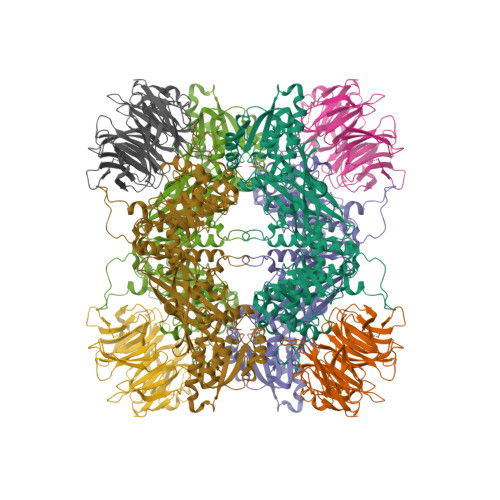
|
|
||||||||||||
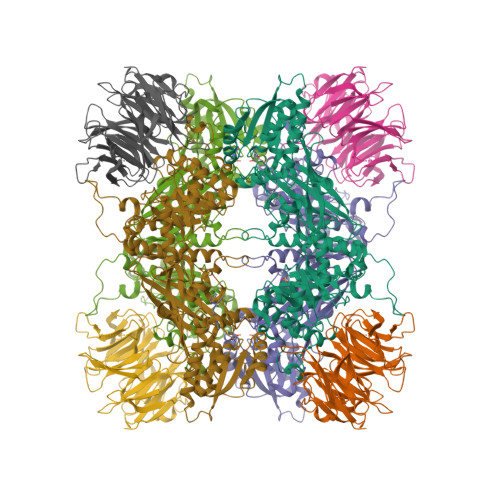
|
|
||||||||||||

|
|
||||||||||||
Download all structure-activity data for this target as a CSV file 
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
1. Alinari L, Mahasenan KV, Yan F, Karkhanis V, Chung JH, Smith EM, Quinion C, Smith PL, Kim L, Patton JT et al.. (2015) Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation. Blood, 125 (16): 2530-43. [PMID:25742700]
2. Brehmer D, Beke L, Wu T, Millar HJ, Moy C, Sun W, Mannens G, Pande V, Boeckx A, van Heerde E et al.. (2021) Discovery and Pharmacological Characterization of JNJ-64619178, a Novel Small-Molecule Inhibitor of PRMT5 with Potent Antitumor Activity. Mol Cancer Ther, 20 (12): 2317-2328. [PMID:34583982]
3. Cottrell KM, Briggs KJ, Whittington DA, Jahic H, Ali JA, Davis CB, Gong S, Gotur D, Gu L, McCarren P et al.. (2024) Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP-Deleted Cancers. J Med Chem, 67 (8): 6064-6080. [PMID:38595098]
4. Jensen-Pergakes K, Tatlock J, Maegley KA, McAlpine IJ, McTigue M, Xie T, Dillon CP, Wang Y, Yamazaki S, Spiegel N et al.. (2022) SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance. Mol Cancer Ther, 21 (1): 3-15. [PMID:34737197]
5. Li QQ, Quan X, Wang ZX, Qiao N, Ni XF, Jing XL, Zhou SS, Tian XL, Zheng GC, Zhan KN et al.. (2025) Design, Synthesis, and Biological Evaluation of 3,4-Dihydroisoquinolin-1(2H)-one Derivatives as Protein Arginine Methyltransferase 5 Inhibitors for the Treatment of Non-Hodgkin's Lymphoma. J Med Chem, 68 (1): 108-134. [PMID:39722476]
6. SGC. GSK591 A chemical probe for PRMT5. Accessed on 11/12/2015. Modified on 04/08/2023. thesgc.org, https://www.thesgc.org/chemical-probes/GSK591
7. SGC. LLY-283 A Chemical Probe For PRMT5. Accessed on 05/04/2017. Modified on 04/08/2023. thesgc.org, https://www.thesgc.org/chemical-probes/LLY-283283
8. Smil D, Eram MS, Li F, Kennedy S, Szewczyk MM, Brown PJ, Barsyte-Lovejoy D, Arrowsmith CH, Vedadi M, Schapira M. (2015) Discovery of a Dual PRMT5-PRMT7 Inhibitor. ACS Med Chem Lett, 6 (4): 408-12. [PMID:25893041]
9. Smith CR, Aranda R, Bobinski TP, Briere DM, Burns AC, Christensen JG, Clarine J, Engstrom LD, Gunn RJ, Ivetac A et al.. (2022) Fragment-Based Discovery of MRTX1719, a Synthetic Lethal Inhibitor of the PRMT5•MTA Complex for the Treatment of MTAP-Deleted Cancers. J Med Chem, 65 (3): 1749-1766. [PMID:35041419]
10. Smith JM, Barlaam B, Beattie D, Bradshaw L, Chan HM, Chiarparin E, Collingwood O, Cooke SL, Cronin A, Cumming I et al.. (2024) Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity. J Med Chem, 67 (16): 13604-13638. [PMID:39080842]
2.1.1.- Protein arginine N-methyltransferases: protein arginine methyltransferase 5 . Last modified on 22/01/2025. Accessed on 02/07/2025. IUPHAR/BPS Guide to PHARMACOLOGY, https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1256.