GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
Voltage-gated potassium channels (Kv): Introduction
Introduction
Voltage-gated potassium channels form a large and diverse family that is evolutionarily conserved. There are 40 human voltage-gated potassium channel genes belonging to 12 subfamilies. These KV channels display broad distributions in the nervous system and other tissues. For excitable cells such as neurons, cardiomyocytes, and muscles, KV channels regulate the waveform and firing pattern of action potentials. KV channels may also regulate the cell volume, proliferation, and migration of a wide range of cell types.
Voltage-gated potassium (KV) channels belong to one of the largest and highly evolutionarily conserved ion channel families [8]. Each KV channel contains four similar or identical pore-forming α-subunits, and it may also contain auxiliary β-subunits that could affect the channel function and/or localization [7,33]. Each pore-forming subunit of KV channels contains six transmembrane segments (S1-S6), with the first four transmembrane segments (S1-S4) constituting the voltage sensor and the last two transmembrane segments flanking a pore loop (S5-P-S6) as the pore domain. In addition to the chromosome location of each KV channel gene in human, mouse and rat, the physiological and pharmacological properties of the channel, and its tissue distribution and pathophysiology have been provided by the Ion Channel Database by the Subcommittee on Voltage-gated potassium channels of the Nomenclature Committee of the International Union of Pharmacology (NC-IUPHAR). This article presents an introduction to the diversity and functions of voltage-gated potassium channels.
Evolutionary conservation of potassium channels
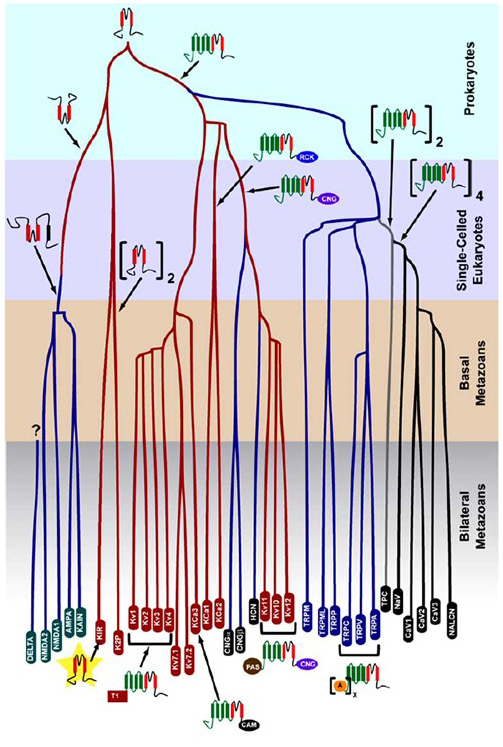
The K+-selectivity that arose in prokaryotes is conserved in a large number of potassium channels with remarkable diversity [14]. As shown in Figure 1, an evolutionary tree of the voltage-gated cation channel superfamily can be proposed based on the comparison of channel genes in the human genome with those in the genomes of other metazoans including mouse, chicken, puffer fish, tunicate, fruit fly, mosquito, nematode and sea anemone [14]. This evolutionary tree envisions a single origin of the K+-selectivity for inwardly rectifying potassium (Kir) channels, voltage-gated potassium (KV) channels, and the two-pore potassium (K2P) channels. It is important to note, however, that a distinct K+-selectivity is likely associated with the evolutionarily conserved organelle K+ channel that resides in endosomes and lysosomes [4].
Figure 1. An evolutionary tree for the genesis of the voltage-gated cation channel superfamily. Based on genome-wide analyses of ion channels from cnidarians and bilateral metazoans [14], this evolutionary tree depicts a common origin for the K+-selectivity of potassium channels (with family names in red ovals), which are related to tetrameric cyclic nucleotide-gated cation (CNG) channels, hyperpolarization-gated cation (HCN) channels and TRP channels, the dimeric TPC channels, and the monomeric Na+, Ca2+, and NALCN channels. The branch lengths do not reflect time. The gene family names at the bottom mark individual branches. Ionotropic glutamate receptors are included based on the hypothesis that they originated from an inversion of the potassium channel pore-forming domain with two transmembrane segments (red). The voltage-sensor domain has four transmembrane segments (green). A: ankryin repeats; CAM: calmodulin-binding domain; CNG: cyclic nucleotide-binding domain; PAS: Per-ARNT-Sim domain; RCK: regulator of conductance of K+ domain; T1: tetramerization domain.
Diversity of voltage-gated potassium channels
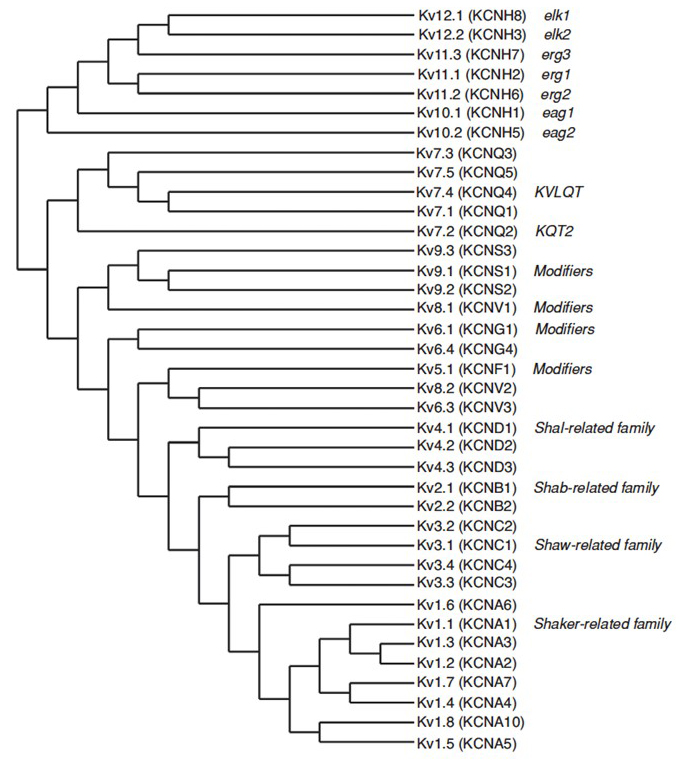
The human genome contains ~80 potassium channel genes of which 40 genes encode voltage-gated potassium channel pore-forming subunits that fall into 12 subfamilies [7] (Fig. 2). Physiologically subdivided into A-type potassium channels that display fast inactivation and delayed rectifier potassium channels without fast inactivation, these KV channels are molecularly and functionally diverse. Fast inactivation, which may impact the action potential duration during repetitive firing, is evident in KV1 channels containing KV1.4 or KVβ1, KV3 channels, and KV4 channels [7]. The delayed rectifier potassium current originally characterized by Hodgkin and Huxley for its role in action potential [9] likely corresponds to the squid KV1 channels [30] that may rely on RNA editing to achieve the flexible functional diversity as many small axons of the giant fiber lobe neurons fuse to form the squid giant axon with greater action potential conduction rate [13,29,36].
Figure 2. Phylogenetic tree for the KV1-12 families. This phylogenetic tree is generated based on analyses of the hydrophobic domain containing the six transmembrane segments (S1-S6) [7]. Both the IUPHAR and the HGNC (in parenthesis) names are shown, along with other commonly used names for these voltage-gated potassium channels.
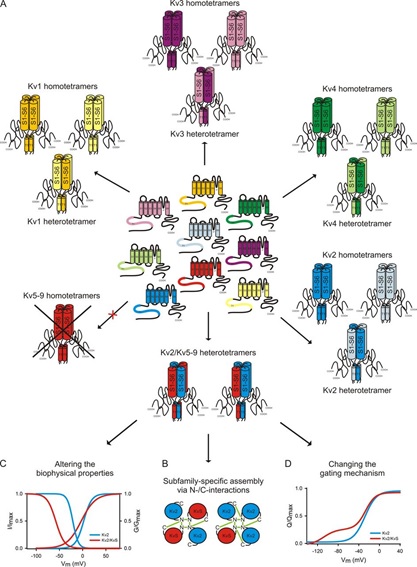
Remarkable diversity of KV channels may be achieved due to the mix and match of KV channel subunits. Within each of the KV1, KV2, KV3, KV4 and KV7 families, homomeric and heteromeric channels may form with a range of functional properties [7,15]. KV2 family members may also assemble with KV5, KV6, KV8 or KV9 family members with more restricted expression patterns in the nervous system and smooth muscles [3], as illustrated schematically in Fig. 3.
Figure 3. KV channel diversity via mix and match of pore-forming channel subunits. (A) The tetrameric KV channels with different properties and distribution encompass homomeric KV1, KV2, KV3, KV4, and KV7 channels, heteromeric channels formed by different members within each of these KV channel families, and heteromeric channels formed by assembly of KV2 family members with KV5, KV6, KV8, or KV9 family members [3]. KV5, KV6, KV8 and KV9 families give rise to homomeric channels that are electrically silent likely due to their retention in the endoplasmic reticulum [3], hence they are referred as KVS. (B) Assembly of KV2 and KVS family members involve their cytoplasmic N- and C-terminal domains. (C, D) Assembly of KV2 and KVS family members gives rise to heteromeric channels with different voltage dependence (C) and gating mechanisms (D) as compared to homomeric channels formed by KV2 family members [3].
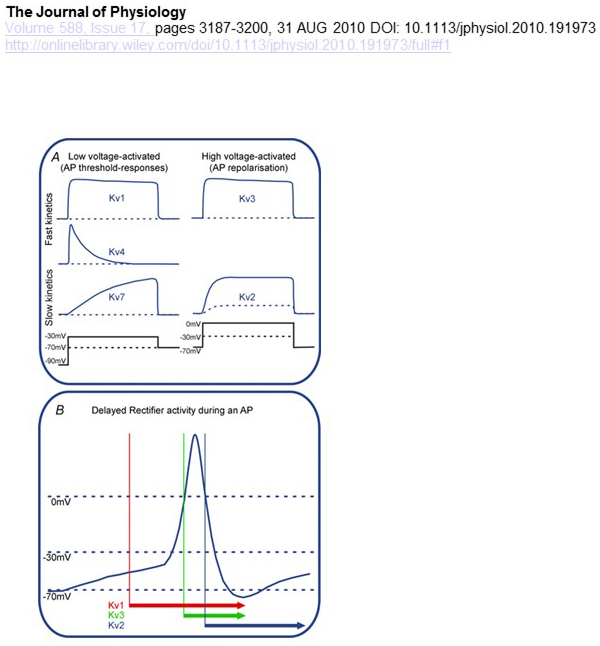
Functional differences in the voltage dependence and kinetics of KV channels underlie their differential contributions to action potential modulation [15] (Fig. 4). Whereas KV1, KV4 and KV7 channels require low levels of membrane depolarization for their activation, KV2 and KV3 channels are activated by greater depolarization. The former, low voltage activated, KV channels may affect the threshold for action potential generation and the number of action potentials generated during depolarization or excitatory synaptic potentials. In contrast, the high voltage activated KV channels may modulate action potential duration and firing pattern [15]. The kinetics of KV channels also influences the ways they contribute to action potential generation. Whereas the low voltage activated KV1 channels with fast activation may affect action potential threshold and waveform, the high voltage activated KV3 channels and KV2 channels can be activated sequentially during an action potential due to the difference in their activation kinetics, and KV2 channels may have more long-lasting effects because of their slow inactivation kinetics. Moreover, KV4 channels with fast inactivation could contribute to the difference in the action potential waveform during repetitive firing, due to suppression of the KV4 channel activity by depolarization [15].
Figure 4. Functional differences of KV channels and their contributions to the action potential. (A) Different KV channels have different voltage dependence for activation and different kinetics [15]. (B) The low voltage activated KV1 channels with fast kinetics open as the cell is depolarized towards the threshold for action potential generation. While both KV2 and KV3 channels are high voltage activated, KV3 channels open sooner than KV2 channels during an action potential. KV2 channels may also take longer to close following an action potential [15].
Potential therapeutic applications of potassium channel modulators
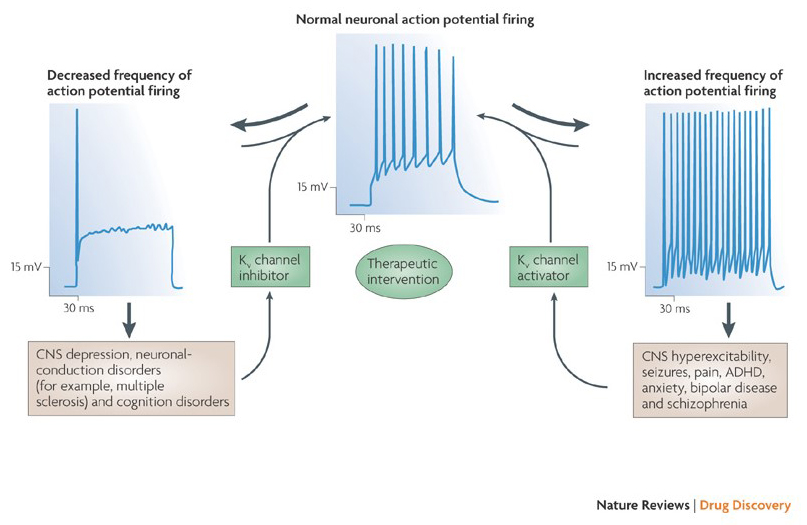
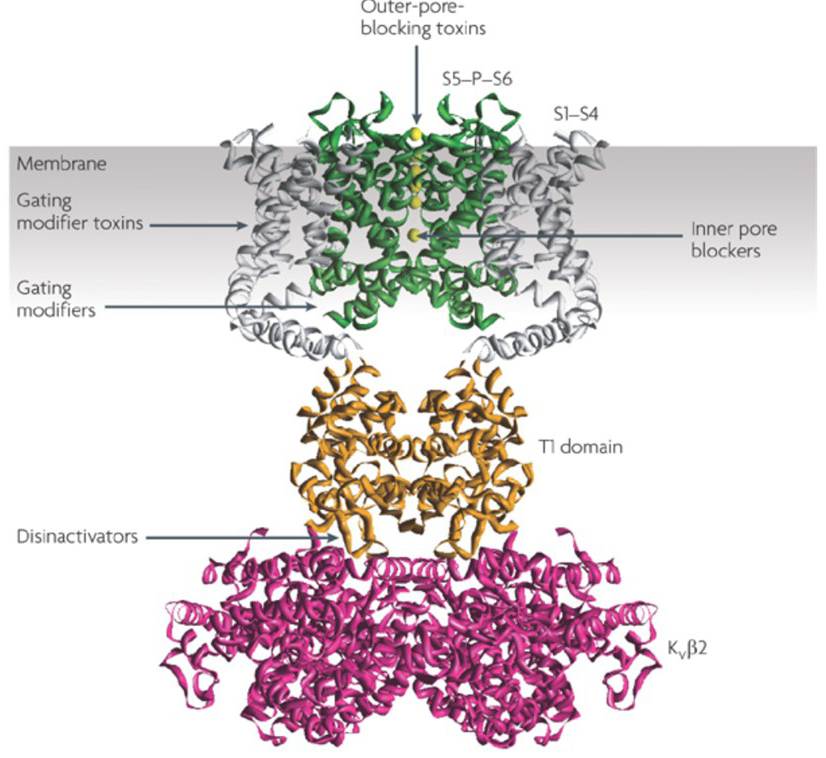
The ability of potassium channel modulators to alter action potential firing patterns has raised the question whether they might be of therapeutic value [35]. As indicated in Fig. 5, various neurological and psychological disorders may involve alterations in the action potential firing patterns, which could be modulated by KV channel activators and blockers [35]. Voltage-gated potassium channels may also play a role in cell proliferation and migration; KV channel modulators have hence been considered for potential treatments of cancer growth and metastasis [2,10-11,18,20,27,32]. In Fig. 6, the KV1.2 channel structure is used schematically to illustrate that KV channel modulators may inhibit channel activity either by occluding the channel permeation pathway, as in the case of outer-pore-blocking toxins and inner pore blockers, or via their interaction with the voltage sensor to stabilized the closed state of the channel, as in the case of gating modifier toxins. Alternatively, some small molecules act by binding to the gating machinery as gating modifiers, or by interacting with the interface between the α- and β-subunits to alter channel activity [35].
Figure 5. Potential applications of KV channel modulators. Because abnormal action potential firing patterns have been associated with diseases such as epilepsy and multiple sclerosis, KV channel activators and inhibitors have been considered for potential therapeutic treatments of diseases that involve alteration of neuronal excitability [35].
Figure 6. Examples of modes of action of KV channel modulators. There are several different ways for peptide toxins and small molecules to modulate KV channel activity. The KV1.2 structure [22] is shown with the pore domains (S5-P-S6) in green, the voltage sensor domains (S1-S4) in grey, the T1 tetramerization domains in orange, and the KVβ2 auxiliary subunits in magenta [35]. Outer-pore-blocking toxins from scorpions, sea anemones, snakes, and cone snails may bind to the outer vestibule and block ion permeation. Gating modifier toxins from spiders such as hanatoxin may interact with the voltage sensor to increase the stability of the closed state, causing rightward shift of the voltage dependence curve for channel activation. There are also small molecule channel modulators that bind to the inner pore (inner pore blockers), the gating hinges (gating modifiers), or the interface between the α- and β-subunits (disinactivators) [35].
Voltage-gated potassium channel structure
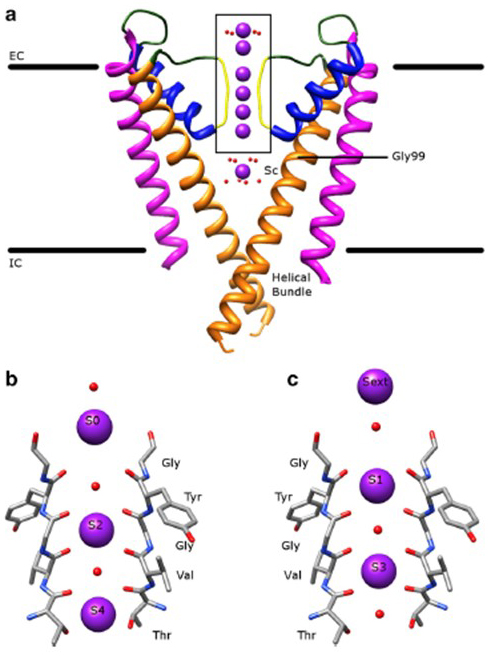
The remarkable selectivity of potassium channels, which allows K+ ions to go through the channel pore with orders of magnitude greater ease than the smaller Na+ ions and with near diffusion-limited rate [8], is accounted for by the ability of the backbone carbonyls of the selectivity filter to coordinate K+ ions that are largely stripped of their hydration shells [25,38], so that more than one K+ ion will move through this narrowest segment of the pore in tandem [16] (Fig. 7) - a long pore for single file K+ ion permeation as predicated [8].
Figure 7. The pore domain of potassium channels. (a) Structure of KcsA in the conductive state (PDB: 1K4C) [38], with the outer helices in magenta, inner helices in orange, pore helices in blue, and the selectivity filter in yellow. K+ ions are in purple while its surrounding water molecules are in red. EC: extracellular; IC: intracellular. (b, c) The selectivity filter in the boxed region of the KcsA structure is shown with K+ ions occupying either the S2 and S4 positions (b) or the S1 and S3 positions (c), to illustrate K+ ion permeation in single file [16].
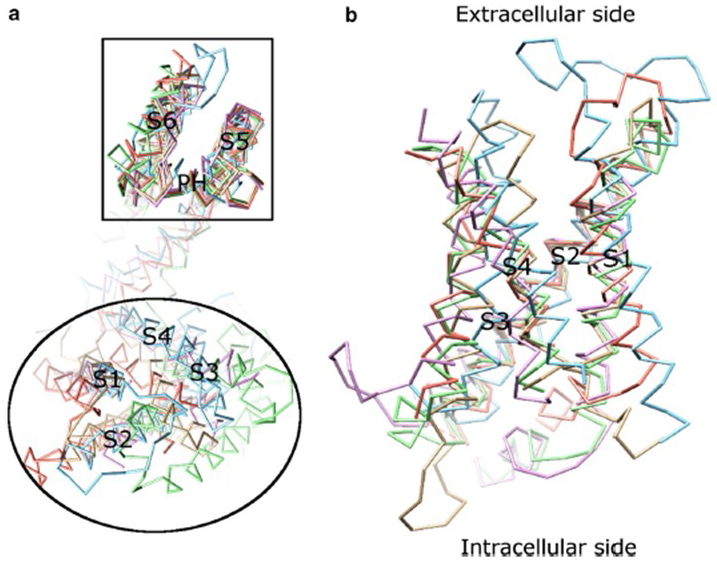
The voltage dependence of KV channel activation [8-9] derives from their voltage sensor domains [12,16,31]. As shown in Fig. 8, KV channels and related channels such as voltage-gated sodium channels and TRPV1 channels in the same superfamily have similar arrangements of their pore domains and voltage sensor domains. The voltage sensor domain of one subunit interacts with the pore domain of a neighboring subunit in a domain swap configuration, and within a voltage sensor the positively charged arginine residues on S4 may interact with negatively charged acidic residues in neighboring helices (Fig. 9).
Figure 8. The voltage sensor domain of voltage-gated potassium channels. (a) Aligning the pore domain (S5-P-S6) of different ion channels reveals that their voltage sensor domains (S1-S4) can take on a variety of orientations (viewed from the extracellular side) [16]. (b) Superimposition of the voltage sensor domain of KV1.2 (PDB: 3LUT, light magenta) [5] with the voltage sensor domains of MlotiK1 (PDB: 3BEH, light brown) [6], NaVAb (PDB: 3RVY, light green) [28], NaVRh (PDB: 4DXW, light orange) [37] and TRPV1 (PDB: 3J5P, light blue) [21] (viewed from the membrane) [16].
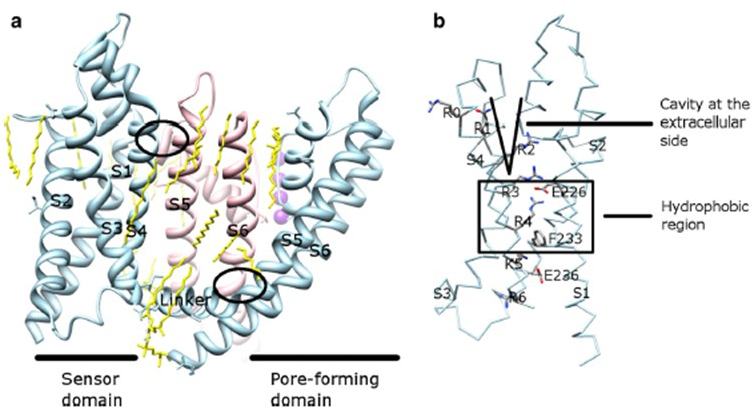
Figure 9. Contacts between the pore domain and the voltage sensor domain of KV channels. (a) The KV1.2-KV2.1 chimera (PDB: 2R9R) [23] with the voltage sensor domain of one subunit (light blue) contacting the pore domain of a neighboring subunit (pink). The contacts on the intracellular side involve the interaction of the S4-S5 linker with S6, and the contacts on the extracellular side involve the interaction between S1 and the pore helix [16]. Lipids (yellow) surrounding the channel and in between the pore domain and the voltage sensor domain are detectable in the crystal structure. (b) Basic residues of S4 and acidic residues in their proximity in the voltage sensor domain [16].
Channelopathies linked to Voltage-gated potassium channels
Voltage-gated potassium channels are broadly expressed in a variety of tissues. In neurons, they are targeted to various subcellular compartments [26,33] (Fig. 10), and channels of different subunit compositions may be present in different subpopulations of neurons [24]. Mutations of KV channel genes may cause neurological diseases such as episodic ataxia and epilepsies, heart diseases, and deafness [1,17,19,34]. Evolutionary conservation of KV channel function is evident, for example, from the similar movement disorders caused by mutation of KV1 orthologs in human, mouse, and the fruit fly [13].
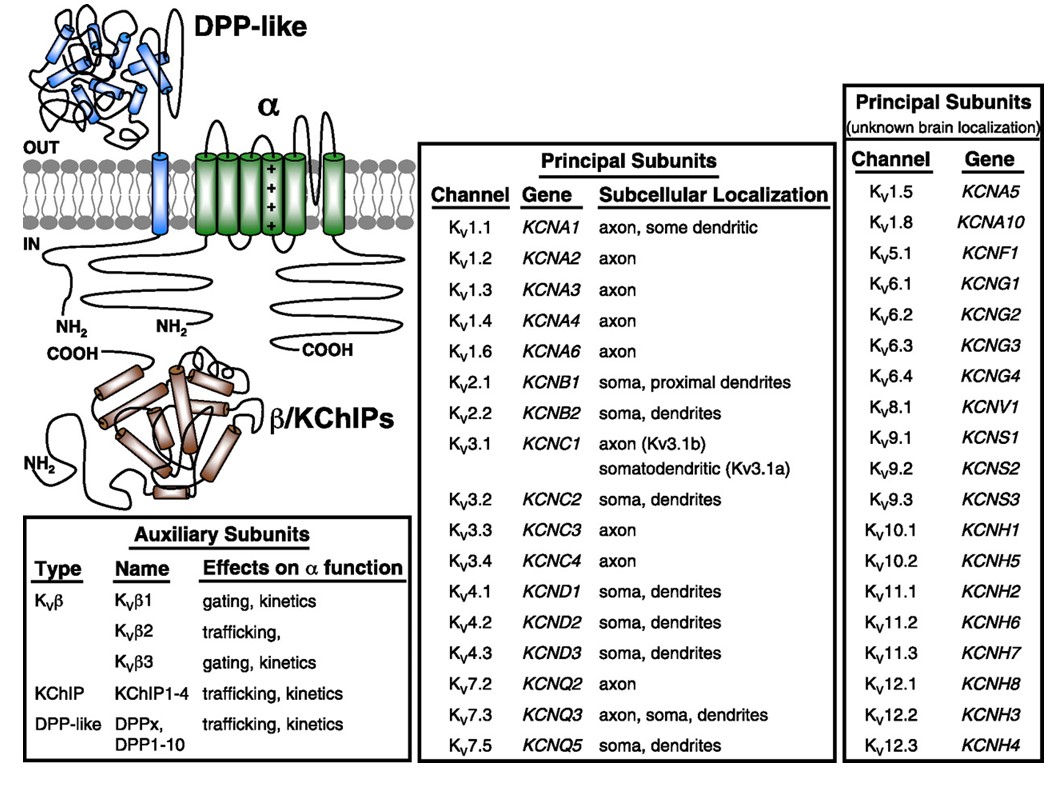
Figure 10. Subcellular distribution of voltage-gated potassium channels. The schematic on the top left depicts a KV4 channel with two different auxiliary subunits. Subcellular localization of various KV channels in mammalian central neurons is indicated in the middle box [33].
References
1. Abriel H, Zaklyazminskaya EV. (2013) Cardiac channelopathies: genetic and molecular mechanisms. Gene, 517 (1): 1-11. [PMID:23266818]
2. Bates E. (2015) Ion channels in development and cancer. Annu Rev Cell Dev Biol, 31: 231-47. [PMID:26566112]
3. Bocksteins E. (2016) Kv5, Kv6, Kv8, and Kv9 subunits: No simple silent bystanders. J Gen Physiol, 147 (2): 105-25. [PMID:26755771]
4. Cang C, Aranda K, Seo YJ, Gasnier B, Ren D. (2015) TMEM175 Is an Organelle K(+) Channel Regulating Lysosomal Function. Cell, 162 (5): 1101-12. [PMID:26317472]
5. Chen X, Wang Q, Ni F, Ma J. (2010) Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement. Proc Natl Acad Sci U S A, 107 (25): 11352-7. [PMID:20534430]
6. Clayton GM, Altieri S, Heginbotham L, Unger VM, Morais-Cabral JH. (2008) Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel. Proc Natl Acad Sci U S A, 105 (5): 1511-5. [PMID:18216238]
7. González C, Baez-Nieto D, Valencia I, Oyarzún I, Rojas P, Naranjo D, Latorre R. (2012) K(+) channels: function-structural overview. Compr Physiol, 2 (3): 2087-149. [PMID:23723034]
8. Hille B. (2001) Ionic Channels of Excitable Membranes, 3rd Ed. In (Sinauer Associates Inc.) .
9. HODGKIN AL, HUXLEY AF. (1952) Currents carried by sodium and potassium ions through the membrane of the giant axon of Loligo. J Physiol, 116 (4): 449-72. [PMID:14946713]
10. Huang X, He Y, Dubuc AM, Hashizume R, Zhang W, Reimand J, Yang H, Wang TA, Stehbens SJ, Younger S et al.. (2015) EAG2 potassium channel with evolutionarily conserved function as a brain tumor target. Nat Neurosci, 18 (9): 1236-46. [PMID:26258683]
11. Huang X, Jan LY. (2014) Targeting potassium channels in cancer. J Cell Biol, 206 (2): 151-62. [PMID:25049269]
12. Isacoff EY, Jan LY, Minor Jr DL. (2013) Conduits of life's spark: a perspective on ion channel research since the birth of neuron. Neuron, 80 (3): 658-74. [PMID:24183018]
13. Jan LY, Jan YN. (2012) Voltage-gated potassium channels and the diversity of electrical signalling. J Physiol, 590 (11): 2591-9. [PMID:22431339]
14. Jegla TJ, Zmasek CM, Batalov S, Nayak SK. (2009) Evolution of the human ion channel set. Comb Chem High Throughput Screen, 12 (1): 2-23. [PMID:19149488]
15. Johnston J, Forsythe ID, Kopp-Scheinpflug C. (2010) Going native: voltage-gated potassium channels controlling neuronal excitability. J Physiol, 588 (Pt 17): 3187-200. [PMID:20519310]
16. Kuang Q, Purhonen P, Hebert H. (2015) Structure of potassium channels. Cell Mol Life Sci, 72 (19): 3677-93. [PMID:26070303]
17. Kullmann DM, Hanna MG. (2002) Neurological disorders caused by inherited ion-channel mutations. Lancet Neurol, 1 (3): 157-66. [PMID:12849484]
18. Kunzelmann K. (2005) Ion channels and cancer. J Membr Biol, 205 (3): 159-73. [PMID:16362504]
19. Lehmann-Horn F, Jurkat-Rott K. (1999) Voltage-gated ion channels and hereditary disease. Physiol Rev, 79 (4): 1317-72. [PMID:10508236]
20. Li M, Xiong ZG. (2011) Ion channels as targets for cancer therapy. Int J Physiol Pathophysiol Pharmacol, 3 (2): 156-66. [PMID:21760973]
21. Liao M, Cao E, Julius D, Cheng Y. (2013) Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature, 504 (7478): 107-12. [PMID:24305160]
22. Long SB, Campbell EB, Mackinnon R. (2005) Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science, 309 (5736): 897-903. [PMID:16002581]
23. Long SB, Tao X, Campbell EB, MacKinnon R. (2007) Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature, 450 (7168): 376-82. [PMID:18004376]
24. Luján R. (2010) Organisation of potassium channels on the neuronal surface. J Chem Neuroanat, 40 (1): 1-20. [PMID:20338235]
25. MacKinnon R. (2003) Potassium channels. FEBS Lett, 555 (1): 62-5. [PMID:14630320]
26. Nusser Z. (2012) Differential subcellular distribution of ion channels and the diversity of neuronal function. Curr Opin Neurobiol, 22 (3): 366-71. [PMID:22033281]
27. Pardo LA. (2004) Voltage-gated potassium channels in cell proliferation. Physiology (Bethesda), 19: 285-92. [PMID:15381757]
28. Payandeh J, Scheuer T, Zheng N, Catterall WA. (2011) The crystal structure of a voltage-gated sodium channel. Nature, 475 (7356): 353-8. [PMID:21743477]
29. Rosenthal JJ, Bezanilla F. (2002) Extensive editing of mRNAs for the squid delayed rectifier K+ channel regulates subunit tetramerization. Neuron, 34 (5): 743-57. [PMID:12062021]
30. Rosenthal JJ, Liu TI, Gilly WF. (1997) A family of delayed rectifier Kv1 cDNAs showing cell type-specific expression in the squid stellate ganglion/giant fiber lobe complex. J Neurosci, 17 (13): 5070-9. [PMID:9185544]
31. Swartz KJ. (2008) Sensing voltage across lipid membranes. Nature, 456 (7224): 891-7. [PMID:19092925]
32. Urrego D, Tomczak AP, Zahed F, Stühmer W, Pardo LA. (2014) Potassium channels in cell cycle and cell proliferation. Philos Trans R Soc Lond B Biol Sci, 369 (1638): 20130094. [PMID:24493742]
33. Vacher H, Mohapatra DP, Trimmer JS. (2008) Localization and targeting of voltage-dependent ion channels in mammalian central neurons. Physiol Rev, 88 (4): 1407-47. [PMID:18923186]
34. Villa C, Combi R. (2016) Potassium Channels and Human Epileptic Phenotypes: An Updated Overview. Front Cell Neurosci, 10: 81. [PMID:27064559]
35. Wulff H, Castle NA, Pardo LA. (2009) Voltage-gated potassium channels as therapeutic targets. Nat Rev Drug Discov, 8 (12): 982-1001. [PMID:19949402]
36. Young JZ. (1939) Fused neurons and synaptic contacts in the giant nerve fibres of cephalopods. Philosophical Transactions of the Royal Society B, (229): 465-503. DOI: 10.1098/rstb.1939.0003
37. Zhang X, Ren W, DeCaen P, Yan C, Tao X, Tang L, Wang J, Hasegawa K, Kumasaka T, He J et al.. (2012) Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature, 486 (7401): 130-4. [PMID:22678295]
38. Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. (2001) Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature, 414 (6859): 43-8. [PMID:11689936]