
GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
Coronavirus (CoV) proteins: Introduction
Figure 1 is a schematic representation of the SARS-CoV-2 RNA genome. The 5′ two-thirds of the genome contains two large, overlapping open reading frames (ORF1 and ORF1b) that encode two long polyprotein precursors, pp1a and pp1ab respectively. These polyproteins are proteolytically cleaved by two integral proteases to generate 14 additional non-structural proteins (16 individual proteins in total). The remaining 3′ third of the genome encodes four structural proteins, spike (S), envelope (E), matrix (M) and nucleocapsid (N), and non-structural proteins, along with a set of accessory proteins. The viral genome hijacks the host cell replication machinery to generate all of the components that are required for self-replication, assembly and release, as well as proteins which manipulate the host's innate immune system. Of the four structural proteins, the RNA winds around the highly basic nucleocapsid (N) protein. The three other structural proteins, E, M and S, are transmembrane proteins. The E protein is a small (9-12 kDa) single transmembrane domain (1TM) protein, which enables virus assembly with the M protein, a larger (23-35 kDa) 3TM protein. Coronaviruses are named for the crown-shaped appearance of the virus due to the large (>120 kDa) S 1TM glycoprotein, which forms extended homotrimers. The SARS-CoV-2 S protein binds to human lung epithelial cells by interacting with the angiotensin-converting enzyme 2 protease on the cell surface [7-8]. This binding, under the influence of the protease TMPRSS2 facilitates viral entry into the host cells and the release of the genome [3].
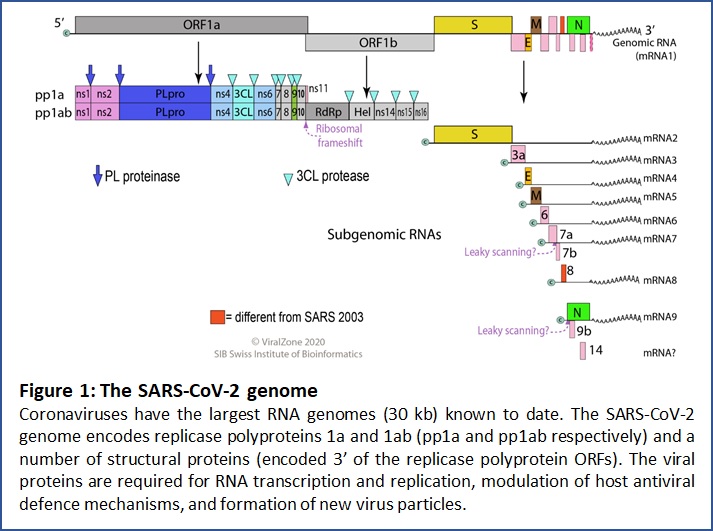
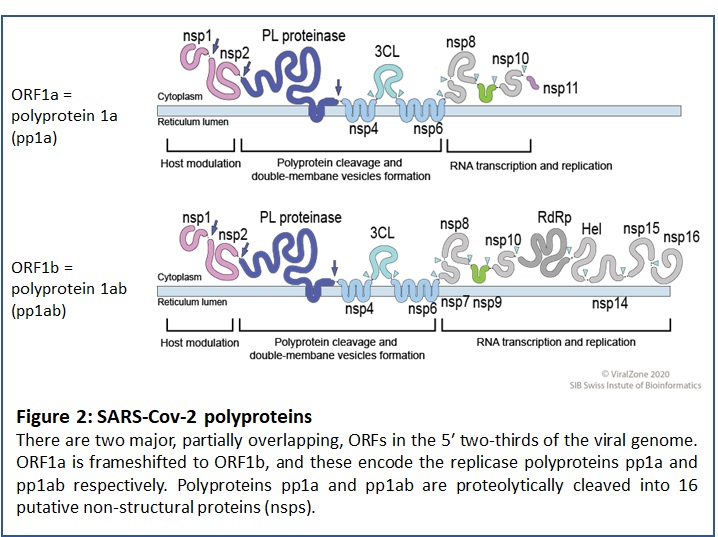
Figure 2 shows the sets of proteins that are generated from the two replicase polyprotein chains that arise from overlapping ORFs at the 5' region of the SARS-CoV-2 RNA. The black arrows indicate PL-pro cleavage sites, and the small light blue arrow heads indicate Mpro cleavage sites within the C-terminal portion of the polyproteins.
In addition to the structural proteins and polyprotein-derived proteins, the SARS-CoV-2 genome encodes 9 accessory factors from a set of sub-genomic open reading frames (ORFs 3, 3a, 6, 7a, 7b, 8, 9b, 9c and 10) within the 3′ region of the genome. SARS-CoV encodes ORF8a and ORF8b, in contrast to SARS-CoV-2's intact ORF8. The functions of the ORF proteins are not fully resolved. One recent development is the identification of deletion mutations in ORF8 of some SARS-CoV-2 isolates [2,6,9]. Evidence from patients with the Δ382 variant suggests that this causes milder infection than the wild type virus [9]. Similar deletion variants in the SARS-CoV ORF8 orthologue were identified [1]. The SARS-CoV Δ29 variant exhibited reduced replication efficiency and was hypothesised to result in a milder clinical illness [4]. In COVID-19 patients the Δ382 variant was associated with reduced systemic release of proinflammatory modulators (cytokines, chemokines, growth factors) that are drivers of severe COVID-19 pathology [9] . In addition, it has been reported that in the Δ382 variant, expression of two viral 'interferon antagonist' genes, ORF6 and N is reduced [2,6], which may offer some explanation as to why this variant seems to be less pathogenic.
Crystallisation and 3D structures of ACE2, Mpro (3CL, nsp5) and S proteins are being generated to provide insight into some of the essential protein-protein interactions and/or protein functions that are required for SARS-CoV-2 infection and replication. To date the leading candidates as novel COVID-19 therapeutic targets that have potential for pharmacological modulation are ACE2 (and its interaction with S; to reduce viral infection) and Mpro (as the key protease for replicase polyprotein processing; to inhibit virus replication).
A dedicated Coronavirus page has been created within the Guide to PHARMACOLOGY, as a portal for the rapid and frequent release of pharmacology-relevent SARS-CoV-2/COVID-19 information. A link to a review article, prepared by IUPHAR and the Guide to PHARMACOLOGY team, which offers insight into potential short- mid- and long-term anti-COVID-19 therapeutic strategies, based on sound pharmacological principles, is included on the Coronavirus page. It is forseeable that like HIV, and in light of the failure to identify monotherapeutics following the original SARS-CoV outbreak in 2003 (despite intense drug discovery activity) [5], effective treament of COVID-19 will rely on a multi-pronged approach that simultaneously targets a number of key elements such as virus replication, its mechanism of infection and its pathological effects in the host.
Figures 1 and 2 are reproduced with permission from the Swiss Bioinformatics Institute.
References
1. Chinese SARS Molecular Epidemiology Consortium. (2004) Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science, 303 (5664): 1666-9. [PMID:14752165]
2. Gong YN, Tsao KC, Hsiao MJ, Huang CG, Huang PN, Huang PW, Lee KM, Liu YC, Yang SL, Kuo RL et al.. (2020) SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg Microbes Infect, 9 (1): 1457-1466. [PMID:32543353]
3. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A et al.. (2020) SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell, 181 (2): 271-280.e8. [PMID:32142651]
4. Muth D, Corman VM, Roth H, Binger T, Dijkman R, Gottula LT, Gloza-Rausch F, Balboni A, Battilani M, Rihtarič D et al.. (2018) Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci Rep, 8 (1): 15177. [PMID:30310104]
5. Pillaiyar T, Manickam M, Namasivayam V, Hayashi Y, Jung SH. (2016) An Overview of Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3CL Protease Inhibitors: Peptidomimetics and Small Molecule Chemotherapy. J Med Chem, 59 (14): 6595-628. [PMID:26878082]
6. Su YCF, Anderson DE, Young BE, Linster M, Zhu F, Jayakumar J, Zhuang Y, Kalimuddin S, Low JGH, Tan CW et al.. (2020) Discovery and Genomic Characterization of a 382-Nucleotide Deletion in ORF7b and ORF8 during the Early Evolution of SARS-CoV-2. mBio, 11 (4). DOI: 10.1128/mBio.01610-20 [PMID:32694143]
7. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. (2020) Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell, 181 (2): 281-292.e6. [PMID:32155444]
8. Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY et al.. (2020) Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell, 181 (4): 894-904.e9. [PMID:32275855]
9. Young BE, Fong S-W, Chan Y-H, Mak T-M, Ang LW, Anderson DE. (2020) Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. The Lancet, [Epub ahead of print]. DOI: 10.1016/S0140-6736(20)31757-8







