|
Compound class:
Natural product
Comment: Darobactin A is an antibacterial compound isolated from Photorhabdus khanii, a species of bacteria that exists symbiotically in the gut of entomopathogenic nematodes [ 1]. Darobactin A has activity against a range of Gram-negative bacteria, including drug-resistant strains of Escherichia coli and Klebsiella pneumoniae [ 1]. It binds to BamA, the central component of the essential bacterial β-barrel assembly machinery (BAM) complex, which facilitates the folding and insertion of β-barrel proteins into the outer membrane of Gram-negative bacteria [ 1-2].
|
|
2D Structure
|
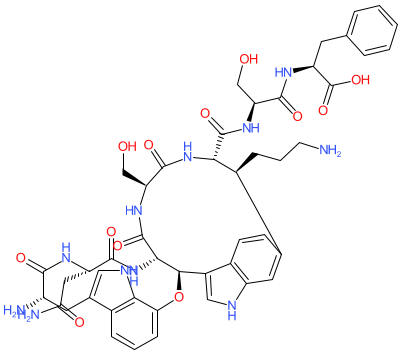
|
Physico-chemical Properties
|
|
|
Hydrogen bond acceptors
|
23
|
|
Hydrogen bond donors
|
14
|
|
Rotatable bonds
|
16
|
|
Topological polar surface area
|
380.78
|
|
Molecular weight
|
966.01
|
|
XLogP
|
-3.7
|
|
No. Lipinski's rules broken
|
4
|
Generated using the Chemistry Development Kit (CDK) (Willighagen EL et al. Journal of Cheminformatics vol. 9:33. 2017, doi:10.1186/s13321-017-0220-4; https://cdk.github.io/)
|
SMILES / InChI / InChIKey
|
|
|
Canonical SMILES
|
C1=CC=C(C=C1)C[C@@H](C(=O)O)NC(=O)[C@H](CO)NC(=O)[C@@H]2[C@@H](CCCN)C3=CC4=C(C=C3)C(=CN4)[C@@H]5[C@@H](C(=O)N[C@@H](CO)C(=O)N2)NC(=O)[C@H](CC(=O)N)NC(=O)[C@H](CC6=CNC7=C6C=CC=C7O5)N
|
|
Isomeric SMILES
|
C1[C@@H](C(=O)N[C@H](C(=O)N[C@H]2[C@@H](C3=CNC4=C3C=CC(=C4)[C@@H]([C@H](NC(=O)[C@@H](NC2=O)CO)C(=O)N[C@@H](CO)C(=O)N[C@@H](CC5=CC=CC=C5)C(=O)O)CCCN)OC6=CC=CC7=C6NC=C71)CC(=O)N)N
|
|
InChI
|
InChI=1S/C47H55N11O12/c48-13-5-9-25-23-11-12-27-28(19-51-30(27)16-23)40-39(58-42(63)31(17-36(50)61)53-41(62)29(49)15-24-18-52-37-26(24)8-4-10-35(37)70-40)46(67)56-34(21-60)44(65)57-38(25)45(66)55-33(20-59)43(64)54-32(47(68)69)14-22-6-2-1-3-7-22/h1-4,6-8,10-12,16,18-19,25,29,31-34,38-40,51-52,59-60H,5,9,13-15,17,20-21,48-49H2,(H2,50,61)(H,53,62)(H,54,64)(H,55,66)(H,56,67)(H,57,65)(H,58,63)(H,68,69)/t25-,29-,31-,32-,33-,34-,38-,39-,40+/m0/s1
|
|
InChI Key
|
HRMCRRLPTVSYHJ-KUDSBEMESA-N
|
Generated using the Chemistry Development Kit (CDK) (Willighagen EL et al. Journal of Cheminformatics vol. 9:33. 2017, doi:10.1186/s13321-017-0220-4; https://cdk.github.io/)
|
|




