
Top ▲

GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
Gene and Protein Information  |
||||||
| class C G protein-coupled receptor | ||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | 7 | 1212 | 11q14.2-q14.3 | GRM5 | glutamate metabotropic receptor 5 | 60-61 |
| Mouse | 7 | 1203 | 7 D3 | Grm5 | glutamate receptor, metabotropic 5 | |
| Rat | 7 | 1203 | 1q32 | Grm5 | glutamate metabotropic receptor 5 | 1,41 |
Previous and Unofficial Names  |
| mGluR5 | GPRC1E | glutamate receptor |
Database Links  |
|
| Specialist databases | |
| GPCRdb | grm5_human (Hs), grm5_mouse (Mm), grm5_rat (Rn) |
| Other databases | |
| Alphafold | P41594 (Hs), Q3UVX5 (Mm), P31424 (Rn) |
| ChEMBL Target | CHEMBL3227 (Hs), CHEMBL1641352 (Mm), CHEMBL2564 (Rn) |
| DrugBank Target | P41594 (Hs) |
| Ensembl Gene | ENSG00000168959 (Hs), ENSMUSG00000049583 (Mm), ENSRNOG00000016429 (Rn) |
| Entrez Gene | 2915 (Hs), 108071 (Mm), 24418 (Rn) |
| Human Protein Atlas | ENSG00000168959 (Hs) |
| KEGG Gene | hsa:2915 (Hs), mmu:108071 (Mm), rno:24418 (Rn) |
| OMIM | 604102 (Hs) |
| Pharos | P41594 (Hs) |
| RefSeq Nucleotide | NM_000842 (Hs), NM_001143831 (Hs), NM_001081414 (Mm), NM_017012 (Rn) |
| RefSeq Protein | NP_001137303 (Hs), NP_000833 (Hs), NP_001074883 (Mm), NP_058708 (Rn) |
| UniProtKB | P41594 (Hs), Q3UVX5 (Mm), P31424 (Rn) |
| Wikipedia | GRM5 (Hs) |
Selected 3D Structures  |
|||||||||||||
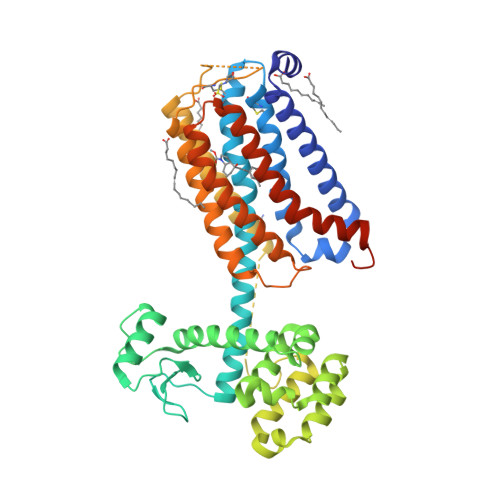
|
|
||||||||||||
Natural/Endogenous Ligands  |
| L-glutamic acid |
| Comments: Other endogenous ligands include L-aspartic acid, L-serine-O-phosphate, NAAG and L-cysteine sulphinic acid |
Download all structure-activity data for this target as a CSV file 
| Agonists | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific agonist tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Agonist Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Indicated affinities were determined by displacement studies of [3H]quisqualate bound on HEK cell membranes expressing a recombinant rat mGlu5. This is the only study describing affinity values for these compounds. More information on agonist potencies determined from functional studies can be obtained from [85]. So far no differences have been reported for the agonist affinities between the rat and the human receptor. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antagonists | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific antagonist tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antagonist Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Indicated affinities were determined by displacement studies of [3H]quisqualate bound on HEK cell membranes expressing a recombinant rat mGlu5 (except for LY341495 value determined from functional studies). This is the only study describing affinity values for these compounds. More information on agonist potencies determined from functional studies can be obtained from [85]. So far no differences have been reported for the antagonist affinities between the rat and the human receptor. Surprisingly, although LY367385 and AIDA displace Quisqualate binding, no antagonistic activity of these compounds have been observed in functional studies (see [85]). (S)-4C3HPG has been reported to have slight partial agonist activity at this receptor [41]. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Allosteric Modulators | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific allosteric modulator tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Allosteric Modulator Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| M-5MPEP is a partal antagonist at the rat receptor [78]. Allosteric regulators, either negative (non-competitive antagonists), positive (allosteric potentiators), or neutral, are usually highly selective for mGlu5, being inactive on the highly homologous mGlu1 receptor. These modulators bind in the 7TM region of the receptor [54,68], whereas the orthosteric site is located in the large extracellular domain of this receptor. A second important point is that many negative modulators possess inverse agonist activity. However, allosteric "partial antagonists" have now been discovered [78]. The concept of partial antagonist activity is novel and can only be achieved with compounds acting at an allosteric site. Although a clear interaction between DFB, CDPPB and the MPEP binding site has been described, the relationship between binding to this site and allosteric potentiation is not yet clear. CPPHA clearly binds at a site different from MPEP [67]. DCB and 5MPEP have been shown to inhibit the effect of both positive and negative allosteric modulators, and represents as such a novel type of modulator called a neutral allosteric site ligand [66,78]. M-5MPEP and Br5-MPEPy fully displace binding of [3H]methoxyPEPy but do not fully block glutamate responses [78]. VU0357121 binds to a site other than the MPEP site [33]. VU0365396 is a non-MPEP site, neutral allosteric ligand [33]. [18F]FPEB has been used in receptor occupancy in humans [96,104]. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Primary Transduction Mechanisms 
|
|
| Transducer | Effector/Response |
| Gq/G11 family | Phospholipase C stimulation |
| Comments: The main action of mGlu5 is to activate PLC via Gq/11. This can be measured via the inositol phosphate production or increase in intracelular Ca2+ both in cell lines expressing this recombinant receptor or in native tissue such as brain slices or cultured neurons or astrocytes. | |
| References: 18,73 | |
Secondary Transduction Mechanisms  |
|
| Transducer | Effector/Response |
|
Gs family Gi/Go family |
Adenylyl cyclase stimulation Potassium channel Calcium channel Phospholipase A2 stimulation Phospholipase D stimulation Other - See Comments |
|
Comments:
Stimulation of adenylyl cyclase has been reported in transfected cell lines, and in native tissue although it is not yet known whether this effect is due to mGlu5 or mGlu1 or both [18]. Coupling of group-I mGlu receptors to PTX-sensitive G-proteins Gi and Go has also been observed in many cells including neurons. Such a coupling is at the origin of MAPK activation. Activation of PLA2 is potentiated by co-activation of the NMDA receptor [24]. PLD activation has been reported both in brain slices [46] and in cultured astrocytes [86]. However, some studies reported this response may also be generated by an mglu receptor with specific pharmacological properties [7,18]. Activation of big potassium currents likely results from the increase in intracellular Ca2+. However, inhibition of various type of potassium currents lead to an increase in cell excitability [4]. Both inhibition and potentiation of voltage sensitive Ca2+ channels (N and L types) have been reported [4,18,73]. |
|
| References: 4,18,73 | |
Tissue Distribution 
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
Expression Datasets  |
|
|
Functional Assays 
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
Physiological Functions 
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
Physiological Consequences of Altering Gene Expression 
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
Phenotypes, Alleles and Disease Models 
|
Mouse data from MGI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Biologically Significant Variants 
|
||||||||
|
||||||||
|
1. Abe T, Sugihara H, Nawa H, Shigemoto R, Mizuno N, Nakanishi S. (1992) Molecular characterization of a novel metabotropic glutamate receptor mGluR5 coupled to inositol phosphate/Ca2+ signal transduction. J Biol Chem, 267: 13361-13368. [PMID:1320017]
2. Amato RJ, Felts AS, Rodriguez AL, Venable DF, Morrison RD, Byers FW, Daniels JS, Niswender CM, Conn PJ, Lindsley CW et al.. (2013) Substituted 1-Phenyl-3-(pyridin-2-yl)urea negative allosteric modulators of mGlu5: discovery of a new tool compound VU0463841 with activity in rat models of cocaine addiction. ACS Chem Neurosci, 4 (8): 1217-28. [PMID:23682684]
3. Anderson JJ, Rao SP, Rowe B, Giracello DR, Holtz G, Chapman DF, Tehrani L, Bradbury MJ, Cosford ND, Varney MA. (2002) [3H]Methoxymethyl-3-[(2-methyl-1,3-thiazol-4-yl)ethynyl]pyridine binding to metabotropic glutamate receptor subtype 5 in rodent brain: in vitro and in vivo characterization. J Pharmacol Exp Ther, 303 (3): 1044-51. [PMID:12438526]
4. Anwyl R. (1999) Metabotropic glutamate receptors: electrophysiological properties and role in plasticity. Brain Res Brain Res Rev, 29 (1): 83-120. [PMID:9974152]
5. Bhave G, Karim F, Carlton SM, Gereau 4th RW. (2001) Peripheral group I metabotropic glutamate receptors modulate nociception in mice. Nat Neurosci, 4 (4): 417-23. [PMID:11276233]
6. Bonnefous C, Vernier JM, Hutchinson JH, Chung J, Reyes-Manalo G, Kamenecka T. (2005) Dipyridyl amides: potent metabotropic glutamate subtype 5 (mGlu5) receptor antagonists. Bioorg Med Chem Lett, 15 (4): 1197-200. [PMID:15686941]
7. Boss V, Nutt KM, Conn PJ. (1994) L-cysteine sulfinic acid as an endogenous agonist of a novel metabotropic receptor coupled to stimulation of phospholipase D activity. Mol Pharmacol, 45: 1177-1182. [PMID:8022410]
8. Brabet I, Parmentier ML, De Colle C, Bockaert J, Acher F, Pin JP. (1998) Comparative effect of L-CCG-I, DCG-IV and gamma-carboxy-L-glutamate on all cloned metabotropic glutamate receptor subtypes. Neuropharmacology, 37 (8): 1043-51. [PMID:9833633]
9. Brice NL, Varadi A, Ashcroft SJ, Molnar E. (2002) Metabotropic glutamate and GABA(B) receptors contribute to the modulation of glucose-stimulated insulin secretion in pancreatic beta cells. Diabetologia, 45 (2): 242-52. [PMID:11935156]
10. Brodkin J, Bradbury M, Busse C, Warren N, Bristow LJ, Varney MA. (2002) Reduced stress-induced hyperthermia in mGluR5 knockout mice. Eur J Neurosci, 16 (11): 2241-4. [PMID:12473093]
11. Brody SA, Dulawa SC, Conquet F, Geyer MA. (2004) Assessment of a prepulse inhibition deficit in a mutant mouse lacking mGlu5 receptors. Mol Psychiatry, 9 (1): 35-41. [PMID:14699440]
12. Burdi DF, Hunt R, Fan L, Hu T, Wang J, Guo Z, Huang Z, Wu C, Hardy L, Detheux M et al.. (2010) Design, synthesis, and structure-activity relationships of novel bicyclic azole-amines as negative allosteric modulators of metabotropic glutamate receptor 5. J Med Chem, 53 (19): 7107-18. [PMID:20809633]
13. Büttelmann B, Peters JU, Ceccarelli S, Kolczewski S, Vieira E, Prinssen EP, Spooren W, Schuler F, Huwyler J, Porter RH et al.. (2006) Arylmethoxypyridines as novel, potent and orally active mGlu5 receptor antagonists. Bioorg Med Chem Lett, 16 (7): 1892-7. [PMID:16439120]
14. Ceccarelli SM, Jaeschke G, Buettelmann B, Huwyler J, Kolczewski S, Peters JU, Prinssen E, Porter R, Spooren W, Vieira E. (2007) Rational design, synthesis, and structure-activity relationship of benzoxazolones: new potent mglu5 receptor antagonists based on the fenobam structure. Bioorg Med Chem Lett, 17 (5): 1302-6. [PMID:17189691]
15. Chae E, Shin YJ, Ryu EJ, Ji MK, Ryune Cho N, Lee KH, Jeong HJ, Kim SJ, Choi Y, Seok Oh K et al.. (2013) Discovery of biological evaluation of pyrazole/imidazole amides as mGlu5 receptor negative allosteric modulators. Bioorg Med Chem Lett, 23 (7): 2134-9. [PMID:23434029]
16. Chen Y, Nong Y, Goudet C, Hemstapat K, de Paulis T, Pin JP, Conn PJ. (2007) Interaction of novel positive allosteric modulators of metabotropic glutamate receptor 5 with the negative allosteric antagonist site is required for potentiation of receptor responses. Mol Pharmacol, 71 (5): 1389-98. [PMID:17303702]
17. Chiamulera C, Epping-Jordan MP, Zocchi A, Marcon C, Cottiny C, Tacconi S, Corsi M, Orzi F, Conquet F. (2001) Reinforcing and locomotor stimulant effects of cocaine are absent in mGluR5 null mutant mice. Nat Neurosci, 4 (9): 873-4. [PMID:11528416]
18. Conn PJ, Pin JP. (1997) Pharmacology and functions of metabotropic glutamate receptors. Annu Rev Pharmacol Toxicol, 37: 205-37. [PMID:9131252]
19. Corti C, Clarkson RW, Crepaldi L, Sala CF, Xuereb JH, Ferraguti F. (2003) Gene structure of the human metabotropic glutamate receptor 5 and functional analysis of its multiple promoters in neuroblastoma and astroglioma cells. J Biol Chem, 278 (35): 33105-19. [PMID:12783878]
20. Cosford ND, Roppe J, Tehrani L, Schweiger EJ, Seiders TJ, Chaudary A, Rao S, Varney MA. (2003) [3H]-methoxymethyl-MTEP and [3H]-methoxy-PEPy: potent and selective radioligands for the metabotropic glutamate subtype 5 (mGlu5) receptor. Bioorg Med Chem Lett, 13 (3): 351-4. [PMID:12565928]
21. De Colle C, Bessis AS, Bockaert J, Acher F, Pin JP. (2000) Pharmacological characterization of the rat metabotropic glutamate receptor type 8a revealed strong similarities and slight differences with the type 4a receptor. Eur J Pharmacol, 394 (1): 17-26. [PMID:10771029]
22. de Paulis T, Hemstapat K, Chen Y, Zhang Y, Saleh S, Alagille D, Baldwin RM, Tamagnan GD, Conn PJ. (2006) Substituent effects of N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamides on positive allosteric modulation of the metabotropic glutamate-5 receptor in rat cortical astrocytes. J Med Chem, 49 (11): 3332-44. [PMID:16722652]
23. Doré AS, Okrasa K, Patel JC, Serrano-Vega M, Bennett K, Cooke RM, Errey JC, Jazayeri A, Khan S, Tehan B et al.. (2014) Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature, 511 (7511): 557-62. [PMID:25042998]
24. Dumuis A, Pin JP, Oomagari K, Sebben M, Bockaert J. (1990) Arachidonic acid released from striatal neurons by joint stimulation of ionotropic and metabotropic quisqualate receptors. Nature, 347 (6289): 182-4. [PMID:1975645]
25. Felts AS, Lindsley SR, Lamb JP, Rodriguez AL, Menon UN, Jadhav S, Jones CK, Conn PJ, Lindsley CW, Emmitte KA. (2010) 3-Cyano-5-fluoro-N-arylbenzamides as negative allosteric modulators of mGlu(5): Identification of easily prepared tool compounds with CNS exposure in rats. Bioorg Med Chem Lett, 20 (15): 4390-4. [PMID:20598884]
26. Frati C, Marchese C, Fisichella G, Copani A, Nasca MR, Storto M, Nicoletti F. (2000) Expression of functional mGlu5 metabotropic glutamate receptors in human melanocytes. J Cell Physiol, 183 (3): 364-72. [PMID:10797311]
27. Galatsis P, Yamagata K, Wendt JA, Connolly CJ, Mickelson JW, Milbank JB, Bove SE, Knauer CS, Brooker RM, Augelli-Szafran CE et al.. (2007) Synthesis and SAR comparison of regioisomeric aryl naphthyridines as potent mGlu5 receptor antagonists. Bioorg Med Chem Lett, 17 (23): 6525-8. [PMID:17936624]
28. Gallagher SM, Daly CA, Bear MF, Huber KM. (2004) Extracellular signal-regulated protein kinase activation is required for metabotropic glutamate receptor-dependent long-term depression in hippocampal area CA1. J Neurosci, 24 (20): 4859-64. [PMID:15152046]
29. Gasparini F, Andres H, Flor PJ, Heinrich M, Inderbitzin W, Lingenhöhl K, Müller H, Munk VC, Omilusik K, Stierlin C et al.. (2002) [(3)H]-M-MPEP, a potent, subtype-selective radioligand for the metabotropic glutamate receptor subtype 5. Bioorg Med Chem Lett, 12 (3): 407-9. [PMID:11814808]
30. Gasparini F, Lingenhöhl K, Stoehr N, Flor PJ, Heinrich M, Vranesic I, Biollaz M, Allgeier H, Heckendorn R, Urwyler S et al.. (1999) 2-Methyl-6-(phenylethynyl)-pyridine (MPEP), a potent, selective and systemically active mGlu5 receptor antagonist. Neuropharmacology, 38 (10): 1493-503. [PMID:10530811]
31. Gilmour G, Broad LM, Wafford KA, Britton T, Colvin EM, Fivush A, Gastambide F, Getman B, Heinz BA, McCarthy AP et al.. (2013) In vitro characterisation of the novel positive allosteric modulators of the mGlu₅ receptor, LSN2463359 and LSN2814617, and their effects on sleep architecture and operant responding in the rat. Neuropharmacology, 64: 224-39. [PMID:22884720]
32. Gregory KJ, Noetzel MJ, Rook JM, Vinson PN, Stauffer SR, Rodriguez AL, Emmitte KA, Zhou Y, Chun AC, Felts AS et al.. (2012) Investigating metabotropic glutamate receptor 5 allosteric modulator cooperativity, affinity, and agonism: enriching structure-function studies and structure-activity relationships. Mol Pharmacol, 82 (5): 860-75. [PMID:22863693]
33. Hammond AS, Rodriguez AL, Townsend SD, Niswender CM, Gregory KJ, Lindsley CW, Conn PJ. (2010) Discovery of a Novel Chemical Class of mGlu(5) Allosteric Ligands with Distinct Modes of Pharmacology. ACS Chem Neurosci, 1 (10): 702-716. [PMID:20981342]
34. Hao J, Dehlinger V, Fivush AM, Rudyk HC, Britton TC, Hollinshead SP, Vokits BP, Clark BP, Henry SS, Massey SM et al.. (2013) Discovery of (1R,2R)-N-(4-(6-isopropylpyridin-2-yl)-3-(2-methyl-2H-indazol-5-yl)isothiazol-5-yl)-2-methylcyclopropanecarboxamide, a potent and orally efficacious mGlu5 receptor negative allosteric modulator. Bioorg Med Chem Lett, 23 (5): 1249-52. [PMID:23374867]
35. Hartveit E, Brandstätter JH, Enz R, Wässle H. (1995) Expression of the mRNA of seven metabotropic glutamate receptors (mGluR1 to 7) in the rat retina. An in situ hybridization study on tissue sections and isolated cells. Eur J Neurosci, 7 (7): 1472-83. [PMID:7551173]
36. Hellyer SD, Albold S, Wang T, Chen ANY, May LT, Leach K, Gregory KJ. (2018) "Selective" Class C G Protein-Coupled Receptor Modulators Are Neutral or Biased mGlu5 Allosteric Ligands. Mol Pharmacol, 93 (5): 504-514. [PMID:29514854]
37. Huang D, Poon SF, Chapman DF, Chung J, Cramer M, Reger TS, Roppe JR, Tehrani L, Cosford ND, Smith ND. (2004) 2-(2-[3-(pyridin-3-yloxy)phenyl]-2H-tetrazol-5-yl) pyridine: a highly potent, orally active, metabotropic glutamate subtype 5 (mGlu5) receptor antagonist. Bioorg Med Chem Lett, 14 (22): 5473-6. [PMID:15482906]
38. Huang Y, Narendran R, Bischoff F, Guo N, Zhu Z, Bae SA, Lesage AS, Laruelle M. (2005) A positron emission tomography radioligand for the in vivo labeling of metabotropic glutamate 1 receptor: (3-ethyl-2-[11C]methyl-6-quinolinyl)(cis- 4-methoxycyclohexyl)methanone. J Med Chem, 48 (16): 5096-9. [PMID:16078827]
39. Hughes ZA, Neal SJ, Smith DL, Sukoff Rizzo SJ, Pulicicchio CM, Lotarski S, Lu S, Dwyer JM, Brennan J, Olsen M et al.. (2013) Negative allosteric modulation of metabotropic glutamate receptor 5 results in broad spectrum activity relevant to treatment resistant depression. Neuropharmacology, 66: 202-14. [PMID:22551786]
40. Jaeschke G, Kolczewski S, Spooren W, Vieira E, Bitter-Stoll N, Boissin P, Borroni E, Büttelmann B, Ceccarelli S, Clemann N et al.. (2015) Metabotropic glutamate receptor 5 negative allosteric modulators: discovery of 2-chloro-4-[1-(4-fluorophenyl)-2,5-dimethyl-1H-imidazol-4-ylethynyl]pyridine (basimglurant, RO4917523), a promising novel medicine for psychiatric diseases. J Med Chem, 58 (3): 1358-71. [PMID:25565255]
41. Joly C, Gomeza J, Brabet I, Curry K, Bockaert J, Pin JP. (1995) Molecular, functional, and pharmacological characterization of the metabotropic glutamate receptor type 5 splice variants: comparison with mGluR1. J Neurosci, 15 (5 Pt 2): 3970-81. [PMID:7751958]
42. Kerner JA, Standaert DG, Penney Jr JB, Young AB, Landwehrmeyer GB. (1997) Expression of group one metabotropic glutamate receptor subunit mRNAs in neurochemically identified neurons in the rat neostriatum, neocortex, and hippocampus. Brain Res Mol Brain Res, 48 (2): 259-69. [PMID:9332723]
43. Kingston AE, Lowndes J, Evans N, Clark B, Tomlinson R, Burnett JP, Mayne NG, Cockerham SL, Lodge D. (1998) Sulphur-containing amino acids are agonists for group 1 metabotropic receptors expressed in clonal RGT cell lines. Neuropharmacology, 37 (3): 277-87. [PMID:9681926]
44. Kingston AE, Ornstein PL, Wright RA, Johnson BG, Mayne NG, Burnett JP, Belagaje R, Wu S, Schoepp DD. (1998) LY341495 is a nanomolar potent and selective antagonist of group II metabotropic glutamate receptors. Neuropharmacology, 37 (1): 1-12. [PMID:9680254]
45. Kinney GG, O'Brien JA, Lemaire W, Burno M, Bickel DJ, Clements MK, Chen TB, Wisnoski DD, Lindsley CW, Tiller PR et al.. (2005) A novel selective positive allosteric modulator of metabotropic glutamate receptor subtype 5 has in vivo activity and antipsychotic-like effects in rat behavioral models. J Pharmacol Exp Ther, 313 (1): 199-206. [PMID:15608073]
46. Klein J, Reymann KG, Riedel G. (1997) Activation of phospholipases C and D by the novel metabotropic glutamate receptor agonist tADA. Neuropharmacology, 36 (2): 261-3. [PMID:9144664]
47. Le Poul E, Bessis AS, Lutgens R, Bonnet B, Rocher JP, Epping-Jordan M, Mutel V. (2005) In vitro pharmacological characterisation of selective mGluR5 positive allosteric modulators. Neuropharmacology, 49 (S1): 252-.
48. Lindemann L, Jaeschke G, Michalon A, Vieira E, Honer M, Spooren W, Porter R, Hartung T, Kolczewski S, Büttelmann B et al.. (2011) CTEP: a novel, potent, long-acting, and orally bioavailable metabotropic glutamate receptor 5 inhibitor. J Pharmacol Exp Ther, 339 (2): 474-86. [PMID:21849627]
49. Lindsley CW, Bates BS, Menon UN, Jadhav SB, Kane AS, Jones CK, Rodriguez AL, Conn PJ, Olsen CM, Winder DG et al.. (2011) (3-Cyano-5-fluorophenyl)biaryl negative allosteric modulators of mGlu(5): Discovery of a new tool compound with activity in the OSS mouse model of addiction. ACS Chem Neurosci, 2 (8): 471-482. [PMID:21927650]
50. Lindsley CW, Wisnoski DD, Leister WH, O'brien JA, Lemaire W, Williams Jr DL, Burno M, Sur C, Kinney GG, Pettibone DJ et al.. (2004) Discovery of positive allosteric modulators for the metabotropic glutamate receptor subtype 5 from a series of N-(1,3-diphenyl-1H- pyrazol-5-yl)benzamides that potentiate receptor function in vivo. J Med Chem, 47 (24): 5825-8. [PMID:15537338]
51. Lu YM, Jia Z, Janus C, Henderson JT, Gerlai R, Wojtowicz JM, Roder JC. (1997) Mice lacking metabotropic glutamate receptor 5 show impaired learning and reduced CA1 long-term potentiation (LTP) but normal CA3 LTP. J Neurosci, 17 (13): 5196-205. [PMID:9185557]
52. Lujan R, Nusser Z, Roberts JD, Shigemoto R, Somogyi P. (1996) Perisynaptic location of metabotropic glutamate receptors mGluR1 and mGluR5 on dendrites and dendritic spines in the rat hippocampus. Eur J Neurosci, 8 (7): 1488-500. [PMID:8758956]
53. Luján R, Roberts JD, Shigemoto R, Ohishi H, Somogyi P. (1997) Differential plasma membrane distribution of metabotropic glutamate receptors mGluR1 alpha, mGluR2 and mGluR5, relative to neurotransmitter release sites. J Chem Neuroanat, 13 (4): 219-41. [PMID:9412905]
54. Malherbe P, Kratochwil N, Zenner MT, Piussi J, Diener C, Kratzeisen C, Fischer C, Porter RH. (2003) Mutational analysis and molecular modeling of the binding pocket of the metabotropic glutamate 5 receptor negative modulator 2-methyl-6-(phenylethynyl)-pyridine. Mol Pharmacol, 64 (4): 823-32. [PMID:14500738]
55. Manka JT, Vinson PN, Gregory KJ, Zhou Y, Williams R, Gogi K, Days E, Jadhav S, Herman EJ, Lavreysen H et al.. (2012) Optimization of an ether series of mGlu5 positive allosteric modulators: molecular determinants of MPEP-site interaction crossover. Bioorg Med Chem Lett, 22 (20): 6481-5. [PMID:22981332]
56. Mannaioni G, Attucci S, Missanelli A, Pellicciari R, Corradetti R, Moroni F. (1999) Biochemical and electrophysiological studies on (S)-(+)-2-(3'-carboxybicyclo(1.1.1)pentyl)-glycine (CBPG), a novel mGlu5 receptor agonist endowed with mGlu1 receptor antagonist activity. Neuropharmacology, 38 (7): 917-26. [PMID:10428410]
57. Mannaioni G, Marino MJ, Valenti O, Traynelis SF, Conn PJ. (2001) Metabotropic glutamate receptors 1 and 5 differentially regulate CA1 pyramidal cell function. J Neurosci, 21 (16): 5925-34. [PMID:11487615]
58. Milbank JB, Knauer CS, Augelli-Szafran CE, Sakkab-Tan AT, Lin KK, Yamagata K, Hoffman JK, Zhuang N, Thomas J, Galatsis P et al.. (2007) Rational design of 7-arylquinolines as non-competitive metabotropic glutamate receptor subtype 5 antagonists. Bioorg Med Chem Lett, 17 (16): 4415-8. [PMID:17590335]
59. Miller S, Romano C, Cotman CW. (1995) Growth factor upregulation of a phosphoinositide-coupled metabotropic glutamate receptor in cortical astrocytes. J Neurosci, 15: 6103-6109. [PMID:7666194]
60. Minakami R, Katsuki F, Sugiyama H. (1993) A variant of metabotropic glutamate receptor subtype 5: an evolutionally conserved insertion with no termination codon. Biochem Biophys Res Commun, 194 (2): 622-7. [PMID:7688218]
61. Minakami R, Katsuki F, Yamamoto T, Nakamura K, Sugiyama H. (1994) Molecular cloning and the functional expression of two isoforms of human metabotropic glutamate receptor subtype 5. Biochem Biophys Res Commun, 199 (3): 1136-43. [PMID:7908515]
62. Mueller R, Dawson ES, Meiler J, Rodriguez AL, Chauder BA, Bates BS, Felts AS, Lamb JP, Menon UN, Jadhav SB et al.. (2012) Discovery of 2-(2-benzoxazoyl amino)-4-aryl-5-cyanopyrimidine as negative allosteric modulators (NAMs) of metabotropic glutamate receptor 5 (mGlu₅): from an artificial neural network virtual screen to an in vivo tool compound. ChemMedChem, 7 (3): 406-14. [PMID:22267125]
63. Mutel V, Ellis GJ, Adam G, Chaboz S, Nilly A, Messer J, Bleuel Z, Metzler V, Malherbe P, Schlaeger EJ et al.. (2000) Characterization of [(3)H]Quisqualate binding to recombinant rat metabotropic glutamate 1a and 5a receptors and to rat and human brain sections. J Neurochem, 75 (6): 2590-601. [PMID:11080213]
64. Noetzel MJ, Gregory KJ, Vinson PN, Manka JT, Stauffer SR, Lindsley CW, Niswender CM, Xiang Z, Conn PJ. (2013) A novel metabotropic glutamate receptor 5 positive allosteric modulator acts at a unique site and confers stimulus bias to mGlu5 signaling. Mol Pharmacol, 83 (4): 835-47. [PMID:23348500]
65. Noetzel MJ, Rook JM, Vinson PN, Cho HP, Days E, Zhou Y, Rodriguez AL, Lavreysen H, Stauffer SR, Niswender CM et al.. (2012) Functional impact of allosteric agonist activity of selective positive allosteric modulators of metabotropic glutamate receptor subtype 5 in regulating central nervous system function. Mol Pharmacol, 81 (2): 120-33. [PMID:22021324]
66. O'Brien JA, Lemaire W, Chen TB, Chang RS, Jacobson MA, Ha SN, Lindsley CW, Schaffhauser HJ, Sur C, Pettibone DJ et al.. (2003) A family of highly selective allosteric modulators of the metabotropic glutamate receptor subtype 5. Mol Pharmacol, 64 (3): 731-40. [PMID:12920211]
67. O'Brien JA, Lemaire W, Wittmann M, Jacobson MA, Ha SN, Wisnoski DD, Lindsley CW, Schaffhauser HJ, Rowe B, Sur C et al.. (2004) A novel selective allosteric modulator potentiates the activity of native metabotropic glutamate receptor subtype 5 in rat forebrain. J Pharmacol Exp Ther, 309 (2): 568-77. [PMID:14747613]
68. Pagano A, Ruegg D, Litschig S, Stoehr N, Stierlin C, Heinrich M, Floersheim P, Prezèau L, Carroll F, Pin JP et al.. (2000) The non-competitive antagonists 2-methyl-6-(phenylethynyl)pyridine and 7-hydroxyiminocyclopropan[b]chromen-1a-carboxylic acid ethyl ester interact with overlapping binding pockets in the transmembrane region of group I metabotropic glutamate receptors. J Biol Chem, 275 (43): 33750-8. [PMID:10934211]
69. Parmentier-Batteur S, Hutson PH, Menzel K, Uslaner JM, Mattson BA, O'Brien JA, Magliaro BC, Forest T, Stump CA, Tynebor RM et al.. (2014) Mechanism based neurotoxicity of mGlu5 positive allosteric modulators--development challenges for a promising novel antipsychotic target. Neuropharmacology, 82: 161-73. [PMID:23291536]
70. Peavy RD, Chang MS, Sanders-Bush E, Conn PJ. (2001) Metabotropic glutamate receptor 5-induced phosphorylation of extracellular signal-regulated kinase in astrocytes depends on transactivation of the epidermal growth factor receptor. J Neurosci, 21 (24): 9619-28. [PMID:11739572]
71. Pilla M, Andreoli M, Tessari M, Delle-Fratte S, Roth A, Butler S, Brown F, Shah P, Bettini E, Cavallini P et al.. (2010) The identification of novel orally active mGluR5 antagonist GSK2210875. Bioorg Med Chem Lett, 20 (24): 7521-4. [PMID:21051228]
72. Pin JP, De Colle C, Bessis AS, Acher F. (1999) New perspectives for the development of selective metabotropic glutamate receptor ligands. Eur J Pharmacol, 375 (1-3): 277-94. [PMID:10443583]
73. Pin JP, Duvoisin R. (1995) The metabotropic glutamate receptors: structure and functions. Neuropharmacology, 34 (1): 1-26. [PMID:7623957]
74. Pittolo S, Gómez-Santacana X, Eckelt K, Rovira X, Dalton J, Goudet C, Pin JP, Llobet A, Giraldo J, Llebaria A et al.. (2014) An allosteric modulator to control endogenous G protein-coupled receptors with light. Nat Chem Biol, 10 (10): 813-5. [PMID:25173999]
75. Porter RH, Jaeschke G, Spooren W, Ballard TM, Büttelmann B, Kolczewski S, Peters JU, Prinssen E, Wichmann J, Vieira E et al.. (2005) Fenobam: a clinically validated nonbenzodiazepine anxiolytic is a potent, selective, and noncompetitive mGlu5 receptor antagonist with inverse agonist activity. J Pharmacol Exp Ther, 315 (2): 711-21. [PMID:16040814]
76. Raboisson P, Breitholtz-Emanuelsson A, Dahllöf H, Edwards L, Heaton WL, Isaac M, Jarvie K, Kers A, Minidis AB, Nordmark A et al.. (2012) Discovery and characterization of AZD9272 and AZD6538-Two novel mGluR5 negative allosteric modulators selected for clinical development. Bioorg Med Chem Lett, 22 (22): 6974-9. [PMID:23046966]
77. Rodriguez AL, Grier MD, Jones CK, Herman EJ, Kane AS, Smith RL, Williams R, Zhou Y, Marlo JE, Days EL et al.. (2010) Discovery of novel allosteric modulators of metabotropic glutamate receptor subtype 5 reveals chemical and functional diversity and in vivo activity in rat behavioral models of anxiolytic and antipsychotic activity. Mol Pharmacol, 78 (6): 1105-23. [PMID:20923853]
78. Rodriguez AL, Nong Y, Sekaran NK, Alagille D, Tamagnan GD, Conn PJ. (2005) A close structural analog of 2-methyl-6-(phenylethynyl)-pyridine acts as a neutral allosteric site ligand on metabotropic glutamate receptor subtype 5 and blocks the effects of multiple allosteric modulators. Mol Pharmacol, 68 (6): 1793-802. [PMID:16155210]
79. Rodriguez-Moreno A, Sistiaga A, Lerma J, Sanchez-Prieto J. (1998) Switch from facilitation to inhibition of excitatory synaptic transmission by group I mGluR desensitization. Neuron, 21: 1477-1486. [PMID:9883739]
80. Romano C, Sesma MA, McDonald CT, O'Malley K, Van den Pol AN, Olney JW. (1995) Distribution of metabotropic glutamate receptor mGluR5 immunoreactivity in rat brain. J Comp Neurol, 355 (3): 455-69. [PMID:7636025]
81. Romano C, van den Pol AN, O'Malley KL. (1996) Enhanced early developmental expression of the metabotropic glutamate receptor mGluR5 in rat brain: protein, mRNA splice variants, and regional distribution. J Comp Neurol, 367 (3): 403-12. [PMID:8698900]
82. Rook JM, Noetzel MJ, Pouliot WA, Bridges TM, Vinson PN, Cho HP, Zhou Y, Gogliotti RD, Manka JT, Gregory KJ et al.. (2013) Unique signaling profiles of positive allosteric modulators of metabotropic glutamate receptor subtype 5 determine differences in in vivo activity. Biol Psychiatry, 73 (6): 501-9. [PMID:23140665]
83. Rook JM, Xiang Z, Lv X, Ghoshal A, Dickerson JW, Bridges TM, Johnson KA, Foster DJ, Gregory KJ, Vinson PN et al.. (2015) Biased mGlu5-Positive Allosteric Modulators Provide In Vivo Efficacy without Potentiating mGlu5 Modulation of NMDAR Currents. Neuron, 86 (4): 1029-1040. [PMID:25937172]
84. Roppe J, Smith ND, Huang D, Tehrani L, Wang B, Anderson J, Brodkin J, Chung J, Jiang X, King C et al.. (2004) Discovery of novel heteroarylazoles that are metabotropic glutamate subtype 5 receptor antagonists with anxiolytic activity. J Med Chem, 47 (19): 4645-8. [PMID:15341479]
85. Schoepp DD, Jane DE, Monn JA. (1999) Pharmacological agents acting at subtypes of metabotropic glutamate receptors. Neuropharmacology, 38 (10): 1431-76. [PMID:10530808]
86. Servitja JM, Masgrau R, Sarri E, Picatoste F. (1999) Group I metabotropic glutamate receptors mediate phospholipase D stimulation in rat cultured astrocytes. J Neurochem, 72 (4): 1441-7. [PMID:10098847]
87. Shigemoto R, Kinoshita A, Wada E, Nomura S, Ohishi H, Takada M, Flor PJ, Neki A, Abe T, Nakanishi S et al.. (1997) Differential presynaptic localization of metabotropic glutamate receptor subtypes in the rat hippocampus. J Neurosci, 17 (19): 7503-22. [PMID:9295396]
88. Shigemoto R, Nomura S, Ohishi H, Sugihara H, Nakanishi S, Mizuno N. (1993) Immunohistochemical localization of a metabotropic glutamate receptor, mGluR5, in the rat brain. Neurosci Lett, 163 (1): 53-7. [PMID:8295733]
89. Siméon FG, Brown AK, Zoghbi SS, Patterson VM, Innis RB, Pike VW. (2007) Synthesis and simple 18F-labeling of 3-fluoro-5-(2-(2-(fluoromethyl)thiazol-4-yl)ethynyl)benzonitrile as a high affinity radioligand for imaging monkey brain metabotropic glutamate subtype-5 receptors with positron emission tomography. J Med Chem, 50 (14): 3256-66. [PMID:17571866]
90. Skerry TM, Genever PG. (2001) Glutamate signalling in non-neuronal tissues. Trends Pharmacol Sci, 22 (4): 174-81. [PMID:11282417]
91. Smith ND, Poon SF, Huang D, Green M, King C, Tehrani L, Roppe JR, Chung J, Chapman DP, Cramer M et al.. (2004) Discovery of highly potent, selective, orally bioavailable, metabotropic glutamate subtype 5 (mGlu5) receptor antagonists devoid of cytochrome P450 1A2 inhibitory activity. Bioorg Med Chem Lett, 14 (22): 5481-4. [PMID:15482908]
92. Spanka C, Glatthar R, Desrayaud S, Fendt M, Orain D, Troxler T, Vranesic I. (2010) Piperidyl amides as novel, potent and orally active mGlu5 receptor antagonists with anxiolytic-like activity. Bioorg Med Chem Lett, 20 (1): 184-8. [PMID:19931453]
93. Spear N, Gadient RA, Wilkins DE, Do M, Smith JS, Zeller KL, Schroeder P, Zhang M, Arora J, Chhajlani V. (2011) Preclinical profile of a novel metabotropic glutamate receptor 5 positive allosteric modulator. Eur J Pharmacol, 659 (2-3): 146-54. [PMID:21335002]
94. Storto M, de Grazia U, Battaglia G, Felli MP, Maroder M, Gulino A, Ragona G, Nicoletti F, Screpanti I, Frati L et al.. (2000) Expression of metabotropic glutamate receptors in murine thymocytes and thymic stromal cells. J Neuroimmunol, 109 (2): 112-20. [PMID:10996213]
95. Storto M, Sallese M, Salvatore L, Poulet R, Condorelli DF, Dell'Albani P, Marcello MF, Romeo R, Piomboni P, Barone N, Nicoletti F, Nicoletti F, De Blasi A. (2001) Expression of metabotropic glutamate receptors in the rat and human testis. J Endocrinol, 170: 71-78. [PMID:11431139]
96. Sullivan JM, Lim K, Labaree D, Lin SF, McCarthy TJ, Seibyl JP, Tamagnan G, Huang Y, Carson RE, Ding YS et al.. (2013) Kinetic analysis of the metabotropic glutamate subtype 5 tracer [(18)F]FPEB in bolus and bolus-plus-constant-infusion studies in humans. J Cereb Blood Flow Metab, 33 (4): 532-41. [PMID:23250105]
97. van den Pol AN, Romano C, Ghosh P. (1995) Metabotropic glutamate receptor mGluR5 subcellular distribution and developmental expression in hypothalamus. J Comp Neurol, 362 (1): 134-50. [PMID:8576426]
98. Varnes JG, Marcus AP, Mauger RC, Throner SR, Hoesch V, King MM, Wang X, Sygowski LA, Spear N, Gadient R et al.. (2011) Discovery of novel positive allosteric modulators of the metabotropic glutamate receptor 5 (mGlu5). Bioorg Med Chem Lett, 21 (5): 1402-6. [PMID:21295468]
99. Varney MA, Cosford ND, Jachec C, Rao SP, Sacaan A, Lin FF, Bleicher L, Santori EM, Flor PJ, Allgeier H et al.. (1999) SIB-1757 and SIB-1893: selective, noncompetitive antagonists of metabotropic glutamate receptor type 5. J Pharmacol Exp Ther, 290 (1): 170-81. [PMID:10381773]
100. Walker K, Reeve A, Bowes M, Winter J, Wotherspoon G, Davis A, Schmid P, Gasparini F, Kuhn R, Urban L. (2001) mGlu5 receptors and nociceptive function II. mGlu5 receptors functionally expressed on peripheral sensory neurones mediate inflammatory hyperalgesia. Neuropharmacology, 40 (1): 10-9. [PMID:11077066]
101. Wang B, Vernier JM, Rao S, Chung J, Anderson JJ, Brodkin JD, Jiang X, Gardner MF, Yang X, Munoz B. (2004) Discovery of novel modulators of metabotropic glutamate receptor subtype-5. Bioorg Med Chem, 12 (1): 17-21. [PMID:14697765]
102. Weiss JM, Jimenez HN, Li G, April M, Uberti MA, Bacolod MD, Brodbeck RM, Doller D. (2011) 6-Aryl-3-pyrrolidinylpyridines as mGlu5 receptor negative allosteric modulators. Bioorg Med Chem Lett, 21 (16): 4891-9. [PMID:21757343]
103. Wendt JA, Deeter SD, Bove SE, Knauer CS, Brooker RM, Augelli-Szafran CE, Schwarz RD, Kinsora JJ, Kilgore KS. (2007) Synthesis and SAR of 2-aryl pyrido[2,3-d]pyrimidines as potent mGlu5 receptor antagonists. Bioorg Med Chem Lett, 17 (19): 5396-9. [PMID:17723296]
104. Wong DF, Waterhouse R, Kuwabara H, Kim J, Brašić JR, Chamroonrat W, Stabins M, Holt DP, Dannals RF, Hamill TG et al.. (2013) 18F-FPEB, a PET radiopharmaceutical for quantifying metabotropic glutamate 5 receptors: a first-in-human study of radiochemical safety, biokinetics, and radiation dosimetry. J Nucl Med, 54 (3): 388-96. [PMID:23404089]
105. Yamaguchi S, Nakanishi S. (1998) Regional expression and regulation of alternative forms of mRNAs derived from two distinct transcription initiation sites of the rat mGluR5 gene. J Neurochem, 71 (1): 60-8. [PMID:9648851]
106. Yatsushiro S, Yamada H, Hayashi M, Tsuboi S, Moriyama Y. (1999) Functional expression of metabotropic glutamate receptor type 5 in rat pinealocytes. Neuroreport, 10 (7): 1599-603. [PMID:10380988]
107. Zhang Y, Rodriguez AL, Conn PJ. (2005) Allosteric potentiators of metabotropic glutamate receptor subtype 5 have differential effects on different signaling pathways in cortical astrocytes. J Pharmacol Exp Ther, 315 (3): 1212-9. [PMID:16135701]
108. Zhou H, Topiol SW, Grenon M, Jimenez HN, Uberti MA, Smith DG, Brodbeck RM, Chandrasena G, Pedersen H, Madsen JC et al.. (2013) Discovery and structure-activity relationship of 1,3-cyclohexyl amide derivatives as novel mGluR5 negative allosteric modulators. Bioorg Med Chem Lett, 23 (5): 1398-406. [PMID:23357634]
109. Zhou Y, Manka JT, Rodriguez AL, Weaver CD, Days EL, Vinson PN, Jadhav S, Hermann EJ, Jones CK, Conn PJ et al.. (2010) Discovery of N-Aryl Piperazines as Selective mGlu(5) Potentiators with Efficacy in a Rodent Model Predictive of Anti-Psychotic Activity. ACS Med Chem Lett, 1 (8): 433-438. [PMID:23308336]
110. Zonta M, Angulo MC, Gobbo S, Rosengarten B, Hossmann KA, Pozzan T, Carmignoto G. (2003) Neuron-to-astrocyte signaling is central to the dynamic control of brain microcirculation. Nat Neurosci, 6: 43-50. [PMID:12469126]