
Top ▲

GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
Gene and Protein Information  |
||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| SARS-CoV-2 | 1 | 1273 | Orf2 | S | Spike glycoprotein | |
| SARS-CoV | 1 | 1255 | Orf2 | S | Spike glycoprotein | |
Previous and Unofficial Names  |
| spike protein |
Database Links  |
|
| ChEMBL Target | CHEMBL4662936 (SARS-CoV-2), CHEMBL4802007 (SARS-CoV) |
| Entrez Gene | 43740568 (SARS-CoV-2), 1489668 (SARS-CoV) |
| RefSeq Protein | YP_009825051 (SARS-CoV), YP_009724390 (SARS-CoV-2) |
| UniProtKB | P0DTC2 (SARS-CoV-2), P59594 (SARS-CoV) |
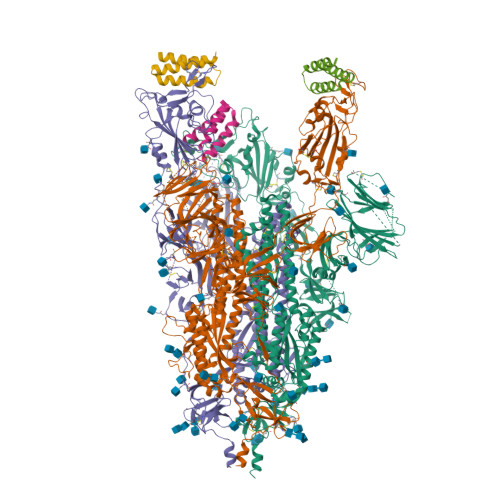
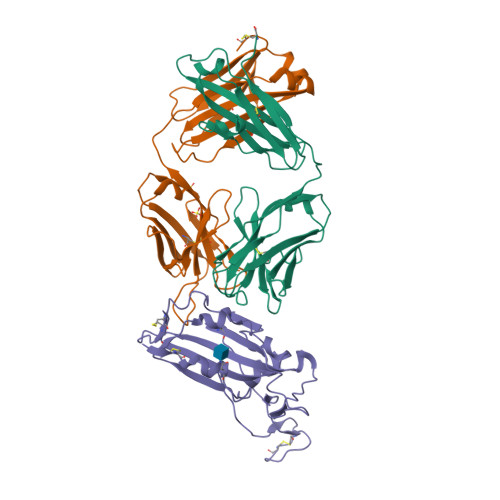
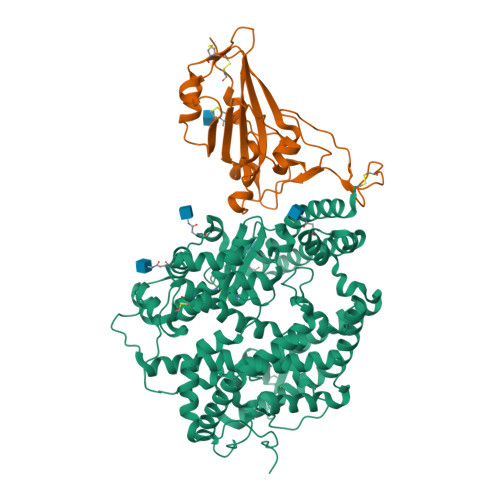
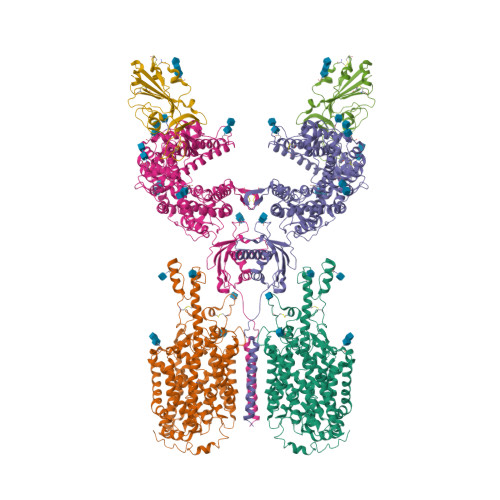
Selected 3D Structures  |
|||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
Download all structure-activity data for this target as a CSV file 
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific inhibitor tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Inhibitor Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EK-1 and EK-1-C4 are viral fusion inhibitors with broad spectrum activity aginst a range of human coronaviruses [20-21], including escape variants [6]. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antibodies | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antibody Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CR3022 binds SARS-CoV-1 spike protein with a Kd of 19 nM, using Spike RBD as analyte in a bio-layer interferometry assay [7]. A number of pharmaceutical companies have anti-SARS-CoV-2 spike biologicals in development. Some companies are concentrating on full construct monoclonal antibodies (mAbs), and others are singling out nanobodies derived from immunised camelids. A selection of the mAbs in development are from Regeneron (REGN10933 and REGN10987, casirivimab and imdevimab respectively; administered as a mixture and granted FDA EUA in late 2020), AstraZeneca (AZD1061 and AZD8895, cilgavimab and tixagevimab respectively), EliLilly (LY3832479, etesevimab and LY-CoV555, bamlanivimab; the latter has FDA EUA), and Celltrion (CT-P59, regdanvimab), Nanobodies are being developed by companies including Dioscure Therapeutics (DIOS-202 and DIOS-203), Twist Bioscience, and University of Texas at Austin/NIH/Ghent University. Adagio Therapeutics have identified an anti-coronavirus mAb (ADG20) with broad-spectrum neutralising activity, including against a number of variants that show varying levels of resistance to existing neutralising antibodies (e.g. P.1 and B.1.351). The epitope for ADG20 resides on the spike protein, at a position that overlaps, but is distinct from the spike's receptor binding domain (RBD: being the target of the majority of existing clinical/lead anti-CoV mAbs). This novel epitope is present in the sarbecoviruses (SARS-CoV and SARS-CoV-2) and some bat coronaviruses, indicating that ADG20 has potential against existing human coronaviruses, and those that may emerge in the future [15]. ADG20 (intramuscular and intravenous doses) will be evaluated in Phase 2/3 clinical trial NCT04805671, in patients with mild-moderate COVID-19 who are at high risk of progressing to severe disease. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Other Binding Ligands | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General Comments |
|
Coronavirus (CoV) spike (S) proteins mediate viral entry by attaching to proteins on target host cells. The SARS coronaviruses exploit ACE2 as their primary entry point. Tor2 was isolated and identified as the prototype SARS-CoV during the extensive SARS outbreak in 2002-2003. Its S protein is highly adapted to bind human ACE2. The SARS CoV S proteins are key to their infection of human cells. S forms trimeric complexes which bind primarily to ACE2 on the surface of host cells to mediate viral entry and to facilitate insertion of its RNA genome into the host cells [24]. Crystal structures of S in closed, open and prefusion configurations have been deposited with the RCSB Protein Data Bank. A structure of spike receptor-binding domain (RBD) bound to ACE2 is represented in PDB accession 6M0J. Experimental evidence suggests that the ACE2 binding domain of SARS-CoV-2 S protein binds to ACE2 more avidly than the homologous domain of SARS-CoV S [18]. The furin cleavage site in SARS-CoV-2's spike protein is necessary for efficient infection, and appears to be required for syncytium formation. Chemical furin inhibitors suppress SARS-CoV-2-induced syncytium and block viral entry and replication [2]. The experimental monoclonal antibody CR3022 was designed to neutralise SARS-CoV [16]. It was developed from an antibody isolated from plasma collected from a convalescent SARS patient. CR3022 was discovered to bind SARS-CoV S protein, including escape mutants. It also binds to the SARS-CoV-2 S protein [25]. X-ray analysis of SARS-CoV-2 spike/CR3022 crystals showed that the antibody targets a highly conserved epitope in S that is distal from the receptor (ACE2)-binding site. The antibody can only gain access to the epitope when at least two RBD on the trimeric S protein are in the "up" conformation. Other anti-spike mAbs were rapidly developed for clinical application. Examples included those from Regeneron (casirivimab + imdevimab) and Eli Lilly (bamlanivimab, etesevimab). The FDA issued Emergency Use Authorisations for the Regeneron cocktail in November 2020, and for single (bamlanivimab) and combined Eli Lilly mAbs (bamlanivimab + etesevimab) in November 2020 and February 2021 respectively. Antibodies that target different components of the virus will potentially be useful as co-agents to anti-spike mAbs for SARS-CoV-2 therapy and/or prophylaxis. A multi-national group of researchers reported on the design and development of spike-based lipopeptides that can prevent spike fusion with host cells [13]. These peptides are based on the heptad repeat (HR) domain at the C terminus of the S protein, and are termed HRC peptides. The most effective construct ([SARSHRC-PEG4]2-chol) was a dimeric PEG-ylated lipopeptide with a cholesterol tag [4] (bioRxiv preprint). Intranasal administration of this lipopeptide completely blocked direct-contact transmission of SARS-CoV-2 in an experimental animal model, and it blocked fusion of SARS-CoV-2 variants (D614G, B.1.1.7 and B.1.351) as well as SARS-CoV and MERS-CoV in VeroE6 cells. These findings support potential of this approach for pre-exposure and early post-exposure prophylaxis of SARS-CoV-2 transmission. |
1. Cao L, Goreshnik I, Coventry B, Case JB, Miller L, Kozodoy L, Chen RE, Carter L, Walls AC, Park YJ et al.. (2020) De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science, 370 (6515): 426-431. [PMID:32907861]
2. Cheng Y-W, Chao T-L, Li C-L, Chen P-J, Chang S-Y, Yeh S-H. (2020) Furin Inhibitors Block SARS-CoV-2 Spike Protein Cleavage to Suppress Virus Production and Cytopathic Effects. Cell Reports, [Epub ahead of print]. DOI: 10.1016/j.celrep.2020.108254
3. Chi Y, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, Zhang Z, Fan P, Dong Y, Yang Y et al.. (2020) A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science,. DOI: 10.1126/science.abc6952
4. de Vries RD, Schmitz KS, Bovier FT, Noack D, Haagmans BL, Biswas S, Rockx B, Gellman SH, Alabi CA, de Swart RL et al.. (2020) Intranasal fusion inhibitory lipopeptide prevents direct contact SARS-CoV-2 transmission in ferrets. bioRxiv, Preprint. DOI: 10.1101/2020.11.04.361154 [PMID:33173865]
5. Hansen J, Baum A, Pascal KE, Russo V, Giordano S, Wloga E, Fulton BO, Yan Y, Koon K, Patel K et al.. (2020) Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science, 369 (6506): 1010-1014. [PMID:32540901]
6. Hoffmann M, Arora P, Groß R, Seidel A, Hörnich BF, Hahn AS, Krüger N, Graichen L, Hofmann-Winkler H, Kempf A et al.. (2021) SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell, [Epub ahead of print]. DOI: 10.1016/j.cell.2021.03.036
7. Huo J, Zhao Y, Ren J, Zhou D, Duyvesteyn HME, Ginn HM, Carrique L, Malinauskas T, Ruza RR, Shah PNM et al.. (2020) Neutralisation of SARS-CoV-2 by destruction of the prefusion Spike. Cell Host and Microbe, In press. DOI: 10.1016/j.chom.2020.06.010
8. Kim C, Ryu DK, Lee J, Kim YI, Seo JM, Kim YG, Jeong JH, Kim M, Kim JI, Kim P et al.. (2021) A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat Commun, 12 (1): 288. [PMID:33436577]
9. Kim SY, Jin W, Sood A, Montgomery DW, Grant OC, Fuster MM, Fu L, Dordick JS, Woods RJ, Zhang F et al.. (2020) Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antiviral Res, 181: 104873. [PMID:32653452]
10. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L et al.. (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature, 581 (7807): 215-220. [PMID:32225176]
11. Loubet P, Gaborit B, Salpin M, Gardeney H, Benotmane I, Systchenko T. (2024) Characteristics of the first immunocompromised patients to receive sipavibart as an early access treatment for COVID-19 pre-exposure prophylaxis in France. Hum Vaccin Immunother, 20 (1): 2387221. [PMID:39143811]
12. Molecular Partners. Molecular Partners pipeline page. Accessed on 16/03/2021. Modified on 16/03/2021. molecularpartners.com, https://www.molecularpartners.com/pipeline/
13. Outlaw VK, Bovier FT, Mears MC, Cajimat MN, Zhu Y, Lin MJ, Addetia A, Lieberman NAP, Peddu V, Xie X et al.. (2020) Inhibition of Coronavirus Entry In Vitro and Ex Vivo by a Lipid-Conjugated Peptide Derived from the SARS-CoV-2 Spike Glycoprotein HRC Domain. mBio, 11 (5). [PMID:33082259]
14. Pei P, Qin H, Chen J, Wang F, He C, He S, Hong B, Liu K, Qiao R, Fan H et al.. (2021) Computational design of ultrashort peptide inhibitors of the receptor-binding domain of the SARS-CoV-2 S protein. Brief Bioinform, 22 (6). [PMID:34180984]
15. Rappazzo CG, Tse LV, Kaku CI, Wrapp D, Sakharkar M, Huang D, Deveau LM, Yockachonis TJ, Herbert AS, Battles MB et al.. (2021) Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science, 371 (6531): 823-829. [PMID:33495307]
16. ter Meulen J, van den Brink EN, Poon LL, Marissen WE, Leung CS, Cox F, Cheung CY, Bakker AQ, Bogaards JA, van Deventer E et al.. (2006) Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med, 3 (7): e237. [PMID:16796401]
17. Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, Lu L, Jiang S, Yang Z, Wu Y et al.. (2020) Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect, 9 (1): 382-385. [PMID:32065055]
18. Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY et al.. (2020) Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell, 181 (4): 894-904.e9. [PMID:32275855]
19. Westendorf K, Wang L, Žentelis S, Foster D, Vaillancourt P, Wiggin M, Lovett E, van der Lee R, Hendle J, Pustilnik A et al.. (2022) LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. bioRxiv, Preprint. DOI: 10.1101/2021.04.30.442182 [PMID:33972947]
20. Xia S, Liu M, Wang C, Xu W, Lan Q, Feng S, Qi F, Bao L, Du L, Liu S et al.. (2020) Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res, 30 (4): 343-355. [PMID:32231345]
21. Xia S, Yan L, Xu W, Agrawal AS, Algaissi A, Tseng CK, Wang Q, Du L, Tan W, Wilson IA et al.. (2019) A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv, 5 (4): eaav4580. [PMID:30989115]
22. Xiang Y, Nambulli S, Xiao Z, Liu H, Sang Z, Duprex WP, Schneidman-Duhovny D, Zhang C, Shi Y. (2020) Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science, 370 (6523): 1479-1484. [PMID:33154108]
23. Yadav PD, Mendiratta SK, Mohandas S, Singh AK, Abraham P, Shete A, Bandyopadhyay S, Kumar S, Parikh A, Kalita P et al.. (2021) ZRC3308 Monoclonal Antibody Cocktail Shows Protective Efficacy in Syrian Hamsters against SARS-CoV-2 Infection. Viruses, 13 (12): 2424. [PMID:34960695]
24. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science, 367 (6485): 1444-1448. [PMID:32132184]
25. Yuan M, Wu NC, Zhu X, Lee CD, So RTY, Lv H, Mok CKP, Wilson IA. (2020) A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science, 368 (6491): 630-633. [PMID:32245784]
26. Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schäfer A, Reidy JX, Trivette A, Nargi RS et al.. (2020) Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature, 584 (7821): 443-449. [PMID:32668443]