
Top ▲

GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
Gene and Protein Information  |
|||||||
| Species | TM | P Loops | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | 6 | 1 | 890 | 5p12 | HCN1 | hyperpolarization activated cyclic nucleotide gated potassium channel 1 | 48 |
| Mouse | 6 | 1 | 910 | 13 66.34 cM | Hcn1 | hyperpolarization activated cyclic nucleotide gated potassium channel 1 | 29,45 |
| Rat | 6 | 1 | 910 | 2q15 | Hcn1 | hyperpolarization-activated cyclic nucleotide-gated potassium channel 1 | 33 |
Database Links  |
|
| Alphafold | O60741 (Hs), O88704 (Mm), Q9JKB0 (Rn) |
| CATH/Gene3D | 2.60.120.10 |
| ChEMBL Target | CHEMBL1795171 (Hs), CHEMBL1250401 (Mm) |
| Ensembl Gene | ENSG00000164588 (Hs), ENSMUSG00000021730 (Mm), ENSRNOG00000055382 (Rn) |
| Entrez Gene | 348980 (Hs), 15165 (Mm), 84390 (Rn) |
| Human Protein Atlas | ENSG00000164588 (Hs) |
| KEGG Gene | hsa:348980 (Hs), mmu:15165 (Mm), rno:84390 (Rn) |
| OMIM | 602780 (Hs) |
| Pharos | O60741 (Hs) |
| RefSeq Nucleotide | NM_021072 (Hs), NM_010408 (Mm), NM_053375 (Rn) |
| RefSeq Protein | NP_066550 (Hs), NP_034538 (Mm), NP_445827 (Rn) |
| SynPHARM | 5496 (in complex with cyclic AMP) |
| UniProtKB | O60741 (Hs), O88704 (Mm), Q9JKB0 (Rn) |
| Wikipedia | HCN1 (Hs) |
Selected 3D Structures  |
|||||||||||||
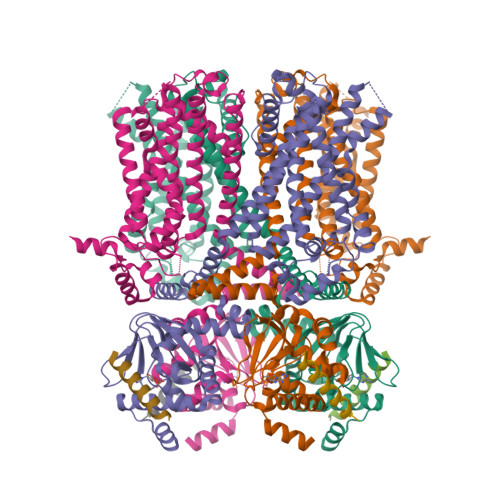
|
|
||||||||||||
Associated Proteins  |
||||||||||||||||||||||||||
|
|
|
||||||||||||||||||||||||
Ion Selectivity and Conductance  |
||||||
|
Voltage Dependence  |
||||||||||||||||||||||
|
||||||||||||||||||||||
|
||||||||||||||||||||||
|
||||||||||||||||||||||
| Activators (Human) |
| cyclic AMP > cyclic GMP (both weak) |
Download all structure-activity data for this target as a CSV file 
| Activators | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Activator Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| cAMP shifts V0.5 by +2 to +7 mV [1,8,53,60]. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Channel Blockers | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific channel blocker tables | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Channel Blocker Comments | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Using HCN1 knockout mice, studies have shown that HCN1 contributes towards the hypnotic actions of the anaesthetics ketamine and isoflurane [9,64]. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tissue Distribution 
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
Physiological Functions 
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
Physiological Consequences of Altering Gene Expression 
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
Phenotypes, Alleles and Disease Models 
|
Mouse data from MGI | ||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||
Clinically-Relevant Mutations and Pathophysiology 
|
||||||||||||||
|
||||||||||||||
|
||||||||||||||
Gene Expression and Pathophysiology 
|
||||||||||||
|
||||||||||||
|
||||||||||||
|
||||||||||||
|
||||||||||||
|
||||||||||||
|
1. Altomare C, Terragni B, Brioschi C, Milanesi R, Pagliuca C, Viscomi C, Moroni A, Baruscotti M, DiFrancesco D. (2003) Heteromeric HCN1-HCN4 channels: a comparison with native pacemaker channels from the rabbit sinoatrial node. J Physiol (Lond.), 549 (Pt 2): 347-59. [PMID:12702747]
2. Bender RA, Soleymani SV, Brewster AL, Nguyen ST, Beck H, Mathern GW, Baram TZ. (2003) Enhanced expression of a specific hyperpolarization-activated cyclic nucleotide-gated cation channel (HCN) in surviving dentate gyrus granule cells of human and experimental epileptic hippocampus. J Neurosci, 23 (17): 6826-36. [PMID:12890777]
3. Bonin RP, Zurek AA, Yu J, Bayliss DA, Orser BA. (2013) Hyperpolarization-activated current (In) is reduced in hippocampal neurons from Gabra5-/- mice. PLoS ONE, 8 (3): e58679. [PMID:23516534]
4. Bucchi A, Tognati A, Milanesi R, Baruscotti M, DiFrancesco D. (2006) Properties of ivabradine-induced block of HCN1 and HCN4 pacemaker channels. J Physiol (Lond.), 572 (Pt 2): 335-46. [PMID:16484306]
5. Budde T, Caputi L, Kanyshkova T, Staak R, Abrahamczik C, Munsch T, Pape HC. (2005) Impaired regulation of thalamic pacemaker channels through an imbalance of subunit expression in absence epilepsy. J Neurosci, 25 (43): 9871-82. [PMID:16251434]
6. Cacheaux LP, Topf N, Tibbs GR, Schaefer UR, Levi R, Harrison NL, Abbott GW, Goldstein PA. (2005) Impairment of hyperpolarization-activated, cyclic nucleotide-gated channel function by the intravenous general anesthetic propofol. J Pharmacol Exp Ther, 315 (2): 517-25. [PMID:16033909]
7. Chan CS, Shigemoto R, Mercer JN, Surmeier DJ. (2004) HCN2 and HCN1 channels govern the regularity of autonomous pacemaking and synaptic resetting in globus pallidus neurons. J Neurosci, 24 (44): 9921-32. [PMID:15525777]
8. Chen S, Wang J, Siegelbaum SA. (2001) Properties of hyperpolarization-activated pacemaker current defined by coassembly of HCN1 and HCN2 subunits and basal modulation by cyclic nucleotide. J Gen Physiol, 117 (5): 491-504. [PMID:11331358]
9. Chen X, Shu S, Kennedy DP, Willcox SC, Bayliss DA. (2009) Subunit-specific effects of isoflurane on neuronal Ih in HCN1 knockout mice. J Neurophysiol, 101 (1): 129-40. [PMID:18971302]
10. Del Lungo M, Melchiorre M, Guandalini L, Sartiani L, Mugelli A, Koncz I, Szel T, Varro A, Romanelli MN, Cerbai E. (2012) Novel blockers of hyperpolarization-activated current with isoform selectivity in recombinant cells and native tissue. Br J Pharmacol, 166 (2): 602-16. [PMID:22091830]
11. Della Santina L, Piano I, Cangiano L, Caputo A, Ludwig A, Cervetto L, Gargini C. (2012) Processing of retinal signals in normal and HCN deficient mice. PLoS ONE, 7 (1): e29812. [PMID:22279546]
12. Du L, Wang SJ, Cui J, He WJ, Ruan HZ. (2013) Inhibition of HCN channels within the periaqueductal gray attenuates neuropathic pain in rats. Behav Neurosci, 127 (2): 325-9. [PMID:23398435]
13. El-Kholy W, MacDonald PE, Fox JM, Bhattacharjee A, Xue T, Gao X, Zhang Y, Stieber J, Li RA, Tsushima RG et al.. (2007) Hyperpolarization-activated cyclic nucleotide-gated channels in pancreatic beta-cells. Mol Endocrinol, 21 (3): 753-64. [PMID:17158221]
14. Giocomo LM, Hussaini SA, Zheng F, Kandel ER, Moser MB, Moser EI. (2011) Grid cells use HCN1 channels for spatial scaling. Cell, 147 (5): 1159-70. [PMID:22100643]
15. Gravante B, Barbuti A, Milanesi R, Zappi I, Viscomi C, DiFrancesco D. (2004) Interaction of the pacemaker channel HCN1 with filamin A. J Biol Chem, 279 (42): 43847-53. [PMID:15292205]
16. Han Y, Noam Y, Lewis AS, Gallagher JJ, Wadman WJ, Baram TZ, Chetkovich DM. (2011) Trafficking and gating of hyperpolarization-activated cyclic nucleotide-gated channels are regulated by interaction with tetratricopeptide repeat-containing Rab8b-interacting protein (TRIP8b) and cyclic AMP at distinct sites. J Biol Chem, 286 (23): 20823-34. [PMID:21504900]
17. He P, Deng J, Zhong X, Zhou Z, Song B, Li L. (2012) Identification of a hyperpolarization-activated cyclic nucleotide-gated channel and its subtypes in the urinary bladder of the rat. Urology, 79 (6): 1411.e7-13. [PMID:22446339]
18. Horwitz GC, Lelli A, Géléoc GS, Holt JR. (2010) HCN channels are not required for mechanotransduction in sensory hair cells of the mouse inner ear. PLoS ONE, 5 (1): e8627. [PMID:20062532]
19. Horwitz GC, Risner-Janiczek JR, Jones SM, Holt JR. (2011) HCN channels expressed in the inner ear are necessary for normal balance function. J Neurosci, 31 (46): 16814-25. [PMID:22090507]
20. Kaupp UB, Seifert R. (2001) Molecular diversity of pacemaker ion channels. Annu Rev Physiol, 63: 235-57. [PMID:11181956]
21. Kim CS, Chang PY, Johnston D. (2012) Enhancement of dorsal hippocampal activity by knockdown of HCN1 channels leads to anxiolytic- and antidepressant-like behaviors. Neuron, 75 (3): 503-16. [PMID:22884333]
22. Knaus A, Zong X, Beetz N, Jahns R, Lohse MJ, Biel M, Hein L. (2007) Direct inhibition of cardiac hyperpolarization-activated cyclic nucleotide-gated pacemaker channels by clonidine. Circulation, 115 (7): 872-80. [PMID:17261653]
23. Koch U, Braun M, Kapfer C, Grothe B. (2004) Distribution of HCN1 and HCN2 in rat auditory brainstem nuclei. Eur J Neurosci, 20 (1): 79-91. [PMID:15245481]
24. Kole MH, Bräuer AU, Stuart GJ. (2007) Inherited cortical HCN1 channel loss amplifies dendritic calcium electrogenesis and burst firing in a rat absence epilepsy model. J Physiol (Lond.), 578 (Pt 2): 507-25. [PMID:17095562]
25. Lee CH, MacKinnon R. (2017) Structures of the Human HCN1 Hyperpolarization-Activated Channel. Cell, 168 (1-2): 111-120.e11. [PMID:28086084]
26. Lewis AS, Schwartz E, Chan CS, Noam Y, Shin M, Wadman WJ, Surmeier DJ, Baram TZ, Macdonald RL, Chetkovich DM. (2009) Alternatively spliced isoforms of TRIP8b differentially control h channel trafficking and function. J Neurosci, 29 (19): 6250-65. [PMID:19439603]
27. Lewis AS, Vaidya SP, Blaiss CA, Liu Z, Stoub TR, Brager DH, Chen X, Bender RA, Estep CM, Popov AB et al.. (2011) Deletion of the hyperpolarization-activated cyclic nucleotide-gated channel auxiliary subunit TRIP8b impairs hippocampal Ih localization and function and promotes antidepressant behavior in mice. J Neurosci, 31 (20): 7424-40. [PMID:21593326]
28. Ludwig A, Zong X, Hofmann F, Biel M. (1999) Structure and function of cardiac pacemaker channels. Cell Physiol Biochem, 9 (4-5): 179-86. [PMID:10575196]
29. Ludwig A, Zong X, Jeglitsch M, Hofmann F, Biel M. (1998) A family of hyperpolarization-activated mammalian cation channels. Nature, 393 (6685): 587-91. [PMID:9634236]
30. Lyashchenko AK, Redd KJ, Yang J, Tibbs GR. (2007) Propofol inhibits HCN1 pacemaker channels by selective association with the closed states of the membrane embedded channel core. J Physiol (Lond.), 583 (Pt 1): 37-56. [PMID:17569731]
31. Michels G, Er F, Khan I, Südkamp M, Herzig S, Hoppe UC. (2005) Single-channel properties support a potential contribution of hyperpolarization-activated cyclic nucleotide-gated channels and If to cardiac arrhythmias. Circulation, 111 (4): 399-404. [PMID:15687126]
32. Mobley AS, Miller AM, Araneda RC, Maurer LR, Müller F, Greer CA. (2010) Hyperpolarization-activated cyclic nucleotide-gated channels in olfactory sensory neurons regulate axon extension and glomerular formation. J Neurosci, 30 (49): 16498-508. [PMID:21147989]
33. Monteggia LM, Eisch AJ, Tang MD, Kaczmarek LK, Nestler EJ. (2000) Cloning and localization of the hyperpolarization-activated cyclic nucleotide-gated channel family in rat brain. Brain Res Mol Brain Res, 81 (1-2): 129-39. [PMID:11000485]
34. Moosmang S, Stieber J, Zong X, Biel M, Hofmann F, Ludwig A. (2001) Cellular expression and functional characterization of four hyperpolarization-activated pacemaker channels in cardiac and neuronal tissues. Eur J Biochem, 268 (6): 1646-52. [PMID:11248683]
35. Much B, Wahl-Schott C, Zong X, Schneider A, Baumann L, Moosmang S, Ludwig A, Biel M. (2003) Role of subunit heteromerization and N-linked glycosylation in the formation of functional hyperpolarization-activated cyclic nucleotide-gated channels. J Biol Chem, 278 (44): 43781-6. [PMID:12928435]
36. Müller F, Scholten A, Ivanova E, Haverkamp S, Kremmer E, Kaupp UB. (2003) HCN channels are expressed differentially in retinal bipolar cells and concentrated at synaptic terminals. Eur J Neurosci, 17 (10): 2084-96. [PMID:12786975]
37. Nolan MF, Dudman JT, Dodson PD, Santoro B. (2007) HCN1 channels control resting and active integrative properties of stellate cells from layer II of the entorhinal cortex. J Neurosci, 27 (46): 12440-51. [PMID:18003822]
38. Nolan MF, Malleret G, Dudman JT, Buhl DL, Santoro B, Gibbs E, Vronskaya S, Buzsáki G, Siegelbaum SA, Kandel ER et al.. (2004) A behavioral role for dendritic integration: HCN1 channels constrain spatial memory and plasticity at inputs to distal dendrites of CA1 pyramidal neurons. Cell, 119 (5): 719-32. [PMID:15550252]
39. Nolan MF, Malleret G, Lee KH, Gibbs E, Dudman JT, Santoro B, Yin D, Thompson RF, Siegelbaum SA, Kandel ER et al.. (2003) The hyperpolarization-activated HCN1 channel is important for motor learning and neuronal integration by cerebellar Purkinje cells. Cell, 115 (5): 551-64. [PMID:14651847]
40. Notomi T, Shigemoto R. (2004) Immunohistochemical localization of Ih channel subunits, HCN1-4, in the rat brain. J Comp Neurol, 471 (3): 241-76. [PMID:14991560]
41. Oh YJ, Na J, Jeong JH, Park DK, Park KH, Ko JS, Kim DS. (2012) Alterations in hyperpolarization-activated cyclic nucleotidegated cation channel (HCN) expression in the hippocampus following pilocarpine-induced status epilepticus. BMB Rep, 45 (11): 635-40. [PMID:23187002]
42. Piskorowski R, Santoro B, Siegelbaum SA. (2011) TRIP8b splice forms act in concert to regulate the localization and expression of HCN1 channels in CA1 pyramidal neurons. Neuron, 70 (3): 495-509. [PMID:21555075]
43. Ramakrishnan NA, Drescher MJ, Barretto RL, Beisel KW, Hatfield JS, Drescher DG. (2009) Calcium-dependent binding of HCN1 channel protein to hair cell stereociliary tip link protein protocadherin 15 CD3. J Biol Chem, 284 (5): 3227-38. [PMID:19008224]
44. Ramakrishnan NA, Drescher MJ, Khan KM, Hatfield JS, Drescher DG. (2012) HCN1 and HCN2 proteins are expressed in cochlear hair cells: HCN1 can form a ternary complex with protocadherin 15 CD3 and F-actin-binding filamin A or can interact with HCN2. J Biol Chem, 287 (45): 37628-46. [PMID:22948144]
45. Santoro B, Grant SG, Bartsch D, Kandel ER. (1997) Interactive cloning with the SH3 domain of N-src identifies a new brain specific ion channel protein, with homology to eag and cyclic nucleotide-gated channels. Proc Natl Acad Sci USA, 94 (26): 14815-20. [PMID:9405696]
46. Santoro B, Hu L, Liu H, Saponaro A, Pian P, Piskorowski RA, Moroni A, Siegelbaum SA. (2011) TRIP8b regulates HCN1 channel trafficking and gating through two distinct C-terminal interaction sites. J Neurosci, 31 (11): 4074-86. [PMID:21411649]
47. Santoro B, Lee JY, Englot DJ, Gildersleeve S, Piskorowski RA, Siegelbaum SA, Winawer MR, Blumenfeld H. (2010) Increased seizure severity and seizure-related death in mice lacking HCN1 channels. Epilepsia, 51 (8): 1624-7. [PMID:20384728]
48. Santoro B, Liu DT, Yao H, Bartsch D, Kandel ER, Siegelbaum SA, Tibbs GR. (1998) Identification of a gene encoding a hyperpolarization-activated pacemaker channel of brain. Cell, 93 (5): 717-29. [PMID:9630217]
49. Santoro B, Piskorowski RA, Pian P, Hu L, Liu H, Siegelbaum SA. (2009) TRIP8b splice variants form a family of auxiliary subunits that regulate gating and trafficking of HCN channels in the brain. Neuron, 62 (6): 802-13. [PMID:19555649]
50. Santoro B, Wainger BJ, Siegelbaum SA. (2004) Regulation of HCN channel surface expression by a novel C-terminal protein-protein interaction. J Neurosci, 24 (47): 10750-62. [PMID:15564593]
51. Seeliger MW, Brombas A, Weiler R, Humphries P, Knop G, Tanimoto N, Müller F. (2011) Modulation of rod photoreceptor output by HCN1 channels is essential for regular mesopic cone vision. Nat Commun, 2: 532. [PMID:22068599]
52. Stevens DR, Seifert R, Bufe B, Müller F, Kremmer E, Gauss R, Meyerhof W, Kaupp UB, Lindemann B. (2001) Hyperpolarization-activated channels HCN1 and HCN4 mediate responses to sour stimuli. Nature, 413 (6856): 631-5. [PMID:11675786]
53. Stieber J, Stöckl G, Herrmann S, Hassfurth B, Hofmann F. (2005) Functional expression of the human HCN3 channel. J Biol Chem, 280 (41): 34635-43. [PMID:16043489]
54. Stieber J, Wieland K, Stöckl G, Ludwig A, Hofmann F. (2006) Bradycardic and proarrhythmic properties of sinus node inhibitors. Mol Pharmacol, 69 (4): 1328-37. [PMID:16387796]
55. Surges R, Kukley M, Brewster A, Rüschenschmidt C, Schramm J, Baram TZ, Beck H, Dietrich D. (2012) Hyperpolarization-activated cation current Ih of dentate gyrus granule cells is upregulated in human and rat temporal lobe epilepsy. Biochem Biophys Res Commun, 420 (1): 156-60. [PMID:22405820]
56. Ulens C, Tytgat J. (2001) Functional heteromerization of HCN1 and HCN2 pacemaker channels. J Biol Chem, 276 (9): 6069-72. [PMID:11133998]
57. Vemana S, Pandey S, Larsson HP. (2008) Intracellular Mg2+ is a voltage-dependent pore blocker of HCN channels. Am J Physiol, Cell Physiol, 295 (2): C557-65. [PMID:18579800]
58. Wainger BJ, DeGennaro M, Santoro B, Siegelbaum SA, Tibbs GR. (2001) Molecular mechanism of cAMP modulation of HCN pacemaker channels. Nature, 411 (6839): 805-10. [PMID:11459060]
59. Wan Y. (2008) Involvement of hyperpolarization-activated, cyclic nucleotide-gated cation channels in dorsal root ganglion in neuropathic pain. Sheng Li Xue Bao, 60 (5): 579-80. [PMID:18958363]
60. Wang J, Chen S, Siegelbaum SA. (2001) Regulation of hyperpolarization-activated HCN channel gating and cAMP modulation due to interactions of COOH terminus and core transmembrane regions. J Gen Physiol, 118 (3): 237-50. [PMID:11524455]
61. Wilkars W, Liu Z, Lewis AS, Stoub TR, Ramos EM, Brandt N, Nicholson DA, Chetkovich DM, Bender RA. (2012) Regulation of axonal HCN1 trafficking in perforant path involves expression of specific TRIP8b isoforms. PLoS ONE, 7 (2): e32181. [PMID:22363812]
62. Yasui K, Liu W, Opthof T, Kada K, Lee JK, Kamiya K, Kodama I. (2001) I(f) current and spontaneous activity in mouse embryonic ventricular myocytes. Circ Res, 88 (5): 536-42. [PMID:11249878]
63. Zhang Y, Liu Y, Qu J, Hardy A, Zhang N, Diao J, Strijbos PJ, Tsushima R, Robinson RB, Gaisano HY et al.. (2009) Functional characterization of hyperpolarization-activated cyclic nucleotide-gated channels in rat pancreatic beta cells. J Endocrinol, 203 (1): 45-53. [PMID:19654142]
64. Zhou C, Douglas JE, Kumar NN, Shu S, Bayliss DA, Chen X. (2013) Forebrain HCN1 channels contribute to hypnotic actions of ketamine. Anesthesiology, 118 (4): 785-95. [PMID:23377220]