
Top ▲

GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
Target not currently curated in GtoImmuPdb
Target id: 626
Nomenclature: Mineralocorticoid receptor
Systematic Nomenclature: NR3C2
Family: 3C. 3-Ketosteroid receptors
Gene and Protein Information  |
|||||
| Species | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | 984 | 4q31 | NR3C2 | nuclear receptor subfamily 3 group C member 2 | 2 |
| Mouse | 978 | 8 36.34 cM | Nr3c2 | nuclear receptor subfamily 3, group C, member 2 | 3 |
| Rat | 981 | 19q11 | Nr3c2 | nuclear receptor subfamily 3, group C, member 2 | 79 |
Previous and Unofficial Names  |
| aldosterone receptor | Type I glucocorticoid receptor | MCR | MLR | MR | nuclear receptor subfamily 3 |
Database Links  |
|
| Specialist databases | |
| Transcriptomine | NR3C2&foldChange=1.6&direction=down, NR3C2&foldChange=1.75&direction=up |
| Other databases | |
| Alphafold | P08235 (Hs), Q8VII8 (Mm), P22199 (Rn) |
| CATH/Gene3D | 3.30.50.10 |
| ChEMBL Target | CHEMBL1994 (Hs), CHEMBL3507 (Rn) |
| DrugBank Target | P08235 (Hs) |
| Ensembl Gene | ENSG00000151623 (Hs), ENSMUSG00000031618 (Mm), ENSRNOG00000034007 (Rn) |
| Entrez Gene | 4306 (Hs), 110784 (Mm), 25672 (Rn) |
| Human Protein Atlas | ENSG00000151623 (Hs) |
| KEGG Gene | hsa:4306 (Hs), mmu:110784 (Mm), rno:25672 (Rn) |
| OMIM | 600983 (Hs) |
| Orphanet | ORPHA123920 (Hs) |
| Pharos | P08235 (Hs) |
| RefSeq Nucleotide | NM_000901 (Hs), NM_001083906 (Mm), NM_013131 (Rn) |
| RefSeq Protein | NP_000892 (Hs), NP_001159576 (Hs), NP_001077375 (Mm), NP_037263 (Rn) |
| SynPHARM |
6580 (in complex with aldosterone) 6578 (in complex with deoxycorticosterone) 6583 (in complex with dexamethasone) 6579 (in complex with progesterone) 78981 (in complex with spironolactone) |
| UniProtKB | P08235 (Hs), Q8VII8 (Mm), P22199 (Rn) |
| Wikipedia | NR3C2 (Hs) |
Selected 3D Structures  |
|||||||||||||
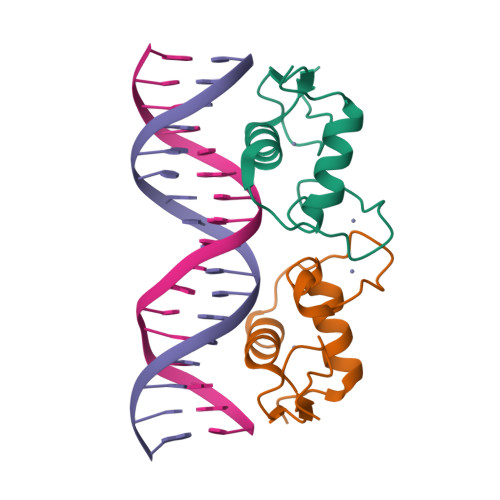
|
|
||||||||||||
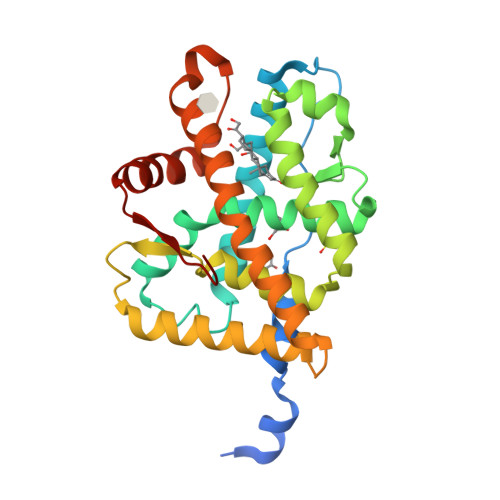
|
|
||||||||||||
Natural/Endogenous Ligands  |
| aldosterone |
| corticosterone |
| cortisol |
| deoxycorticosterone |
| progesterone |
| Rank order of potency (Human) |
| corticosterone, cortisol, aldosterone, progesterone [95] |
Download all structure-activity data for this target as a CSV file 
Agonists  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific agonist tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Antagonists  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antagonist Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Several other antagonists are in various stages of development and are reviewed by Collin et al., 2014 [25]. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
DNA Binding  |
|||||||
|
|||||||
| DNA Binding Comments | |||||||
| MR and GR can heterodimerize. HRE sequence has variations that contribute to gene-specific regulation. Additional response elements are described in Ziera et al., 2009 [128]. Analysis of the cistromes for the human and rat MRs is reported by Le Billan et al. (2015) [61] and van Weert et al. (2017) [114] respectively. |
|||||||
Co-binding Partners  |
|||
| Name | Interaction | Effect | Reference |
| Rac1 | 100 | ||
| HMGD | Physical, Functional | DNA binding | 11,116 |
| Glucocorticoid receptor | Physical, Functional | DNA binding | 63,66-68,75,111 |
| HSP90 complex | Physical, Functional | Cellular localization | 34,38-40,84 |
| Epidermal growth factor receptor | 44 | ||
| Co-binding Partners Comments | |||
| MR interacts with other members of the HSP90 complex including, hsp70, p23, the FκBPs and the cyclopholins [16,85-86]. | |||
Main Co-regulators  |
||||||
| Name | Activity | Specific | Ligand dependent | AF-2 dependent | Comments | References |
| PPARGC1A | Co-activator | No | Yes | No | Strong MR coactivator and highly expressed in brown adipocytes. | 37 |
| PPARGC1A | Co-activator | Yes | Yes | No | Elongation factor that directly interacts with the N-terminal domain of MR and acts as a potent coactivator; strongly represses GR transactivation and has no effect on AR or PR activity. | 78 |
| NCOA1 | Co-activator | No | Yes | Yes | First member of a large coactivator family (SRC1, 2, 3). DNA-bound steroid receptors interact with SRC-1 which initiate sequential recruitment of SWI/SNF chromatin remodeling complexes, histone-methyltransferase proteins CARM1/PRMT1 and histone acetylases such as CBP/p300-pCAF. Sites of interaction with the MR: AF-2; AF-1 by SRC-1e isoform. Recruits histone acetylation complex to initiate transcription; weak intrinsic histone acetyltransferase activity. |
50,76,118,125 |
| NCOA2 | Co-activator | - | No | Yes | Sites of interaction with the MR: AF-1, AF-2. Enhances transactivation. | 37,48,118 |
| EP300 | Co-activator | - | No | Yes | Sites of interaction with the MR: AF-1, AF-2. Exerts histone acetyltransferase activity; recruits RNA polymerase II to target gene promoter. | 37 |
| PPARGC1A | Co-activator | - | No | Yes | Sites of interaction with the MR: AF-2. Recruits histone acetyltransferase complex; facilitates binding of NR to transcription initiation complex. | 50,55 |
| ELL | Co-activator | - | No | - | Sites of interaction with the MR: AF-1b. RNA polymerase II elongation factor; prevents premature arrest and transient pausing of polymerase II. | 78 |
| CASP8AP2 | Co-activator | - | No | - | Sites of interaction with the MR: AF-1. Regulates cell apoptosis. | 72 |
| FAF1 | Co-activator | - | No | - | Sites of interaction with the MR: AF-1. Regulates cell apoptosis. | 72 |
| NRIP1 | Co-activator | - | No | - | Sites of interaction with the MR: N-terminal domain | 125 |
| NCOR2 | Co-repressor | No | No | Yes | Recruited to antagonist bound steroid receptors followed by recruitment of histone deacetylase proteins (HDAC). Sites of interaction with the MR: ligand binding domain. |
118 |
| NCOR1 | Co-repressor | No | No | Yes | Recruited to antagonist bound steroid receptors followed by recruitment of histone deacetylase proteins (HDAC). Sites of interaction with the MR: ligand binding domain. |
118 |
| NFYC | Co-repressor | - | No | - | Sites of interaction with the MR: AF-1. Inhibits aldosterone-induced MR N-C interaction. | 65 |
| PIAS1 | Co-repressor | Yes | Yes | No | PIAS1, a SUMO-E3 ligase, inhibits transactivation by MR and AR but not that by GR. Sites of interaction with the MR: N-terminal domain, possibly ligand binding domain. Exact mechanism of repressive action unclear. | 109 |
| DAXX | Co-repressor | - | No | - | Sites of interaction with the MR: N-terminal domain. Regulates cell apoptosis; represses MR transactivation in some cell lines. | 72 |
| UBE2I | Co-activator | - | No | - | Sites of interaction with the MR: N-terminal domain. SUMO E2-conjugating enzyme; forms coactivation complex with SRC-1. | 122 |
| TRIM24 | Co-activator | - | No | - | Sites of interaction with the MR: N-terminal domain. Transcriptional coactivator / corepressor. | 125 |
| RACK1 | Co-activator | No | Yes | - | The mechanisms of the direct interaction has not been shown. The coactivation is dependent on phosphorylation of RACK1 by PKCβ. | 59 |
| GEMIN4 | Co-repressor | Yes | Yes | Yes | Cell-specific repression of MR signalling. | 120 |
| TESMIN | Co-activator | Yes | Yes | Yes | Sites of interaction with the MR: AF-2. Aldosterone-specific. | 93 |
| Main Co-regulators Comments | ||||||
| A publication describing RHA as a MR co-regulator (Kitagawa et al., 2002) has been retracted [54,69]. XRCC6, EEF1A1 and SSRP1 are non-specific, ligand-dependent coregulators that confer tissue specific regulation [121]. |
||||||
Main Target Genes  |
|||||
| Name | Species | Effect | Technique | Comments | References |
| Sgk1 | Rat | Activated | the focal induction of serum and glucocorticoid-regulated kinase 1 (SGK1) is in the distal nephron and colon | 7,14,101 | |
| Fxyd4 | Rat | Activated | Fxyd4 or the chanel-inducing factor (Chif) is a member of the FXYD membrane protein family associated with Na+K+ATPase. | 13 | |
| K-Ras2A | None | Activated | Northern blot analysis, real-time RT-PCR | Using the Xenopus laevis kidney-derived A6 cell line, the K-ras transcript of the K-ras gene was identified as aldosterone induced. | 15,106 |
| Mmp12 | Mouse | Activated | PCR | Validated as apart of myeloid cell inflammatory response to MR activation. | 74,99 |
| Per1 | Mouse | Activated | Microarray, Northern blot | 45 | |
| Per2 | Mouse | Activated | Microarray, Northern blot | 45 | |
| Ctgf | Mouse | Activated | Microarray, Northern blot | 45 | |
| Cnksr3/CNKSR3 | Mouse | Activated | ChIP | NB: mouse AND human. Validated as part of the regulatory complex associated with ENaC | 103,128 |
| Fkbp5 | Rat | Activated | qRT-PCR of endogenous target gene, Western blotting | 80 | |
| WNK1 | Human | Activated | Semiquantitative RT-PCR | 70 | |
| ICAM1 | Human | Activated | qRT-PCR, immunoblotting | 19 | |
| Edn1 | Rat | Activated | Northern blot analysis, real-time RT-PCR | 119 | |
| Serpine1 | Rat | Activated | RT-PCR | 123 | |
| Ndrg2 | Rat | Activated | RT-PCR | 12 | |
| SCNN1A | Human | Activated | ENaC (SCNN1A) is transcriptionally regulated by aldosterone as an early event in distal colon but not in the kidney. However, aldosterone does increase ENaC number and activity on kidney epithelial cell surface. | 1,102 | |
| GILZ/Tsc22d3 | Mouse | Activated | Serial analysis of gene expression (SAGE) | Subsequently has been validated as a functionally relevant MR regulated gene in several tissues and species. | 87,92,104 |
| Main Target Genes Comments | |||||
| K-Ras2 gene is activated by MR in Xenopus [115]. A small G protein and a proto-oncogene was found to be rapidly induced by aldosterone, enhances Na+ current. Other genes activated include the following: Na+, K+ ATPase α1 and β1 [56-57,57]. In addition to the genes listed above, regulation of L-type Ca2+ channel [62], osteogenic genes including alkaline phosphatase (ALP) and bone morphogenetic protein-2 (BMP2) [52], the RNA polymerase II elongation factor ELL (eleven-nineteen lysine-rich leukemia; [78]), the ubiquitin-specific protease Usp2-45 [33], and Sgk1, Fkbp5, Rasl12, Tns1 and Tsc22d3 (Gilz) which were validated as direct target genes of MR by quantitative RT-qPCR and ChIP-qPCR in a study using a murine distal convoluted tubular epithelial cell-line [112]. |
|||||
Tissue Distribution 
|
||||||||
|
||||||||
| Tissue Distribution Comments | ||||||||
| Classic aldosterone-sensitive tissues include epithelia with high electrical resistance, such as the distal parts of the nephron, the surface epithelium of the distal colon, and salivary and sweat gland ducts. More recently, other MR-expressing cells have been identified, either epithelial, as in epidermal keratinocytes, or nonepithelial, as in the neurons of the central nervous system, the cardiac myocytes, and the endothelial and smooth muscle cells of the vasculature (large vessels). Similar patterns of expression are also seen in rodents. There is also extensive documentation of MR expression in renal and cardiac cell types (reviewed in Odermatt and Kratschmar, 2012 [73]). In addition to those human tissues listed above, there are now many more documented in the literature, including female reproductive tissues (ovary, breast), auditory and retinal tissues, inflammatory cells, particularly the monocyte/macrophage lineage, the rest of the gastrointestinal tract although highest levels are undoubtedly in the distal colon. The MR is co-expressed with 11βHSD2 in a small number of nuclei in the nucleus of the solitary tract (NTS); in these nuclei the role of the MR is to regulate salt appetite in response to aldosterone and therefore sodium balance. They occupy a subregion of the NTS with a diminished blood brain barrier which may afford exposure to circulating aldosterone. These neurons express the angiotensin receptor and MR activation appears to interact synergistically with AngII to promote salt appetite [41,88]. Expression datasets are available from these referenes: [45,83,92,107]. |
||||||||
Functional Assays 
|
||||||||||
|
||||||||||
|
Physiological Consequences of Altering Gene Expression 
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
| Physiological Consequences of Altering Gene Expression Comments | ||||||||||
| There are now a series of tissue-specific MR knockouts. The physiology of these at baseline is limited although in a pathophysiology context the effects are profound (see [24]). | ||||||||||
Phenotypes, Alleles and Disease Models 
|
Mouse data from MGI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Clinically-Relevant Mutations and Pathophysiology 
|
||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||
Biologically Significant Variants 
|
||||||||||||
|
||||||||||||
|
||||||||||||
|
||||||||||||
|
||||||||||||
|
||||||||||||
|
||||||||||||
| Biologically Significant Variant Comments | ||||||||||||
| Human mineralocorticoid receptor isoform 1 (MR-A) has higher transactivation activity than MR-B. Other splicing variants include: A 12-bp insertion at the 3' of exon 3 results in a four-residue addition in between the two zinc fingers of the DBD and no difference in activity compared to the wild-type receptor; a 10-bp deletion in rat MR results truncated LBD at residue 807, unresponsive to aldosterone, and no interference with wild-tpe receptor function; exon-skipping in human generates mutants lacking exon 5 or both exons 5 and 6 which binds to DNA and modulate wild-type receptor activity in a ligand-independent manner [10,125,127]. |
1. Amasheh S, Epple HJ, Mankertz J, Detjen K, Goltz M, Schulzke JD, Fromm M. (2000) Differential regulation of ENaC by aldosterone in rat early and late distal colon. Ann N Y Acad Sci, 915: 92-4. [PMID:11193605]
2. Arriza JL, Weinberger C, Cerelli G, Glaser TM, Handelin BL, Housman DE, Evans RM. (1987) Cloning of human mineralocorticoid receptor complementary DNA: structural and functional kinship with the glucocorticoid receptor. Science, 237 (4812): 268-75. [PMID:3037703]
3. Beggah AT, Escoubet B, Puttini S, Cailmail S, Delage V, Ouvrard-Pascaud A, Bocchi B, Peuchmaur M, Delcayre C, Farman N, Jaisser F. (2002) Reversible cardiac fibrosis and heart failure induced by conditional expression of an antisense mRNA of the mineralocorticoid receptor in cardiomyocytes. Proc Natl Acad Sci USA, 99 (10): 7160-5. [PMID:11997477]
4. Bell MG, Gernert DL, Grese TA, Belvo MD, Borromeo PS, Kelley SA, Kennedy JH, Kolis SP, Lander PA, Richey R et al.. (2007) (S)-N-{3-[1-cyclopropyl-1-(2,4-difluoro-phenyl)-ethyl]-1H-indol-7-yl}-methanesulfonamide: a potent, nonsteroidal, functional antagonist of the mineralocorticoid receptor. J Med Chem, 50 (26): 6443-5. [PMID:18038968]
5. Berger S, Bleich M, Schmid W, Cole TJ, Peters J, Watanabe H, Kriz W, Warth R, Greger R, Schütz G. (1998) Mineralocorticoid receptor knockout mice: pathophysiology of Na+ metabolism. Proc Natl Acad Sci USA, 95 (16): 9424-9. [PMID:9689096]
6. Berger S, Wolfer DP, Selbach O, Alter H, Erdmann G, Reichardt HM, Chepkova AN, Welzl H, Haas HL, Lipp HP et al.. (2006) Loss of the limbic mineralocorticoid receptor impairs behavioral plasticity. Proc Natl Acad Sci U S A, 103 (1): 195-200. [PMID:16368758]
7. Bhargava A, Fullerton MJ, Myles K, Purdy TM, Funder JW, Pearce D, Cole TJ. (2001) The serum- and glucocorticoid-induced kinase is a physiological mediator of aldosterone action. Endocrinology, 142 (4): 1587-94. [PMID:11250940]
8. Bigas J, Sevilla LM, Pérez P. (2020) Epidermal Mineralocorticoid Receptor Inactivation Affects the Homeostasis of All Skin Layers in Chronologically Aged Mice. J Invest Dermatol, 140 (10): 1899-1908. [PMID:32199993]
9. Bledsoe RK, Madauss KP, Holt JA, Apolito CJ, Lambert MH, Pearce KH, Stanley TB, Stewart EL, Trump RP, Willson TM, Williams SP. (2005) A ligand-mediated hydrogen bond network required for the activation of the mineralocorticoid receptor. J Biol Chem, 280 (35): 31283-93. [PMID:15967794]
10. Bloem LJ, Guo C, Pratt JH. (1995) Identification of a splice variant of the rat and human mineralocorticoid receptor genes. J Steroid Biochem Mol Biol, 55 (2): 159-62. [PMID:7495694]
11. Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi ME, Taraseviciene L, Nordeen SK, Allegretto EA, Edwards DP. (1998) High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol, 18 (8): 4471-87. [PMID:9671457]
12. Boulkroun S, Fay M, Zennaro MC, Escoubet B, Jaisser F, Blot-Chabaud M, Farman N, Courtois-Coutry N. (2002) Characterization of rat NDRG2 (N-Myc downstream regulated gene 2), a novel early mineralocorticoid-specific induced gene. J Biol Chem, 277 (35): 31506-15. [PMID:12072429]
13. Brennan FE, Fuller PJ. (1999) Acute regulation by corticosteroids of channel-inducing factor gene messenger ribonucleic acid in the distal colon. Endocrinology, 140 (3): 1213-8. [PMID:10067846]
14. Brennan FE, Fuller PJ. (2000) Rapid upregulation of serum and glucocorticoid-regulated kinase (sgk) gene expression by corticosteroids in vivo. Mol Cell Endocrinol, 166 (2): 129-36. [PMID:10996431]
15. Brennan FE, Fuller PJ. (2006) Mammalian K-ras2 is a corticosteroid-induced gene in vivo. Endocrinology, 147 (6): 2809-16. [PMID:16543373]
16. Bruner KL, Derfoul A, Robertson NM, Guerriero G, Fernandes-Alnemri T, Alnemri ES, Litwack G. (1997) The unliganded mineralocorticoid receptor is associated with heat shock proteins 70 and 90 and the immunophilin FKBP-52. Recept Signal Transduct, 7 (2): 85-98. [PMID:9392437]
17. Bärfacker L, Kuhl A, Hillisch A, Grosser R, Figueroa-Pérez S, Heckroth H, Nitsche A, Ergüden JK, Gielen-Haertwig H, Schlemmer KH et al.. (2012) Discovery of BAY 94-8862: a nonsteroidal antagonist of the mineralocorticoid receptor for the treatment of cardiorenal diseases. ChemMedChem, 7 (8): 1385-403. [PMID:22791416]
18. Canonica J, Sergi C, Maillard M, Klusonova P, Odermatt A, Koesters R, Loffing-Cueni D, Loffing J, Rossier B, Frateschi S et al.. (2016) Adult nephron-specific MR-deficient mice develop a severe renal PHA-1 phenotype. Pflugers Arch, 468 (5): 895-908. [PMID:26762397]
19. Caprio M, Newfell BG, la Sala A, Baur W, Fabbri A, Rosano G, Mendelsohn ME, Jaffe IZ. (2008) Functional mineralocorticoid receptors in human vascular endothelial cells regulate intercellular adhesion molecule-1 expression and promote leukocyte adhesion. Circ Res, 102 (11): 1359-67. [PMID:18467630]
20. Casimiro-Garcia A, Piotrowski DW, Ambler C, Arhancet GB, Banker ME, Banks T, Boustany-Kari CM, Cai C, Chen X, Eudy R et al.. (2014) Identification of (R)-6-(1-(4-cyano-3-methylphenyl)-5-cyclopentyl-4,5-dihydro-1H-pyrazol-3-yl)-2-methoxynicotinic acid, a highly potent and selective nonsteroidal mineralocorticoid receptor antagonist. J Med Chem, 57 (10): 4273-88. [PMID:24738581]
21. Chow CP, Liu JR, Tan XJ, Huang ZH. (2017) Pharmacological Profile of KBP-5074, a Novel NonSteroidal Mineralocorticoid Receptor Antagonist for the Treatment of Cardiorenal Diseases. ournal of Drug Research and Development, 3 (2). DOI: 10.16966/2470-1009.137
22. Coghlan MJ, Kym PR, Elmore SW, Wang AX, Luly JR, Wilcox D, Stashko M, Lin CW, Miner J, Tyree C et al.. (2001) Synthesis and characterization of non-steroidal ligands for the glucocorticoid receptor: selective quinoline derivatives with prednisolone-equivalent functional activity. J Med Chem, 44 (18): 2879-85. [PMID:11520196]
23. Cole TJ, Terella L, Morgan J, Alexiadis M, Yao YZ, Enriori P, Young MJ, Fuller PJ. (2015) Aldosterone-Mediated Renal Sodium Transport Requires Intact Mineralocorticoid Receptor DNA-Binding in the Mouse. Endocrinology, 156 (8): 2958-68. [PMID:26066075]
24. Cole TJ, Young MJ. (2017) 30 YEARS OF THE MINERALOCORTICOID RECEPTOR: Mineralocorticoid receptor null mice: informing cell-type-specific roles. J Endocrinol, 234 (1): T83-T92. [PMID:28550025]
25. Collin M, Niemann F, Jaisser F. (2014) Mineralocorticoid receptor modulators: a patent review (2007 - 2012). Expert Opin Ther Pat, 24 (2): 177-83. [PMID:24215301]
26. Cukier HN, Griswold AJ, Hofmann NK, Gomez L, Whitehead PL, Abramson RK, Gilbert JR, Cuccaro ML, Dykxhoorn DM, Pericak-Vance MA. (2020) Three Brothers With Autism Carry a Stop-Gain Mutation in the HPA-Axis Gene NR3C2. Autism Res, 13 (4): 523-531. [PMID:32064789]
27. de Kloet ER, Van Acker SA, Sibug RM, Oitzl MS, Meijer OC, Rahmouni K, de Jong W. (2000) Brain mineralocorticoid receptors and centrally regulated functions. Kidney Int, 57 (4): 1329-36. [PMID:10760063]
28. De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S et al.. (2014) Synaptic, transcriptional and chromatin genes disrupted in autism. Nature, 515 (7526): 209-15. [PMID:25363760]
29. DeRijk RH, Wüst S, Meijer OC, Zennaro MC, Federenko IS, Hellhammer DH, Giacchetti G, Vreugdenhil E, Zitman FG, de Kloet ER. (2006) A common polymorphism in the mineralocorticoid receptor modulates stress responsiveness. J Clin Endocrinol Metab, 91 (12): 5083-9. [PMID:17018659]
30. Dietz JD, Du S, Bolten CW, Payne MA, Xia C, Blinn JR, Funder JW, Hu X. (2008) A number of marketed dihydropyridine calcium channel blockers have mineralocorticoid receptor antagonist activity. Hypertension, 51 (3): 742-8. [PMID:18250364]
31. Edelman JL, Nehme A. (2013) PHARMACEUTICAL COMPOSITIONS AND METHODS OF USE OF 4-PREGENEN-11ß-17-21-TRIOL-3,20-DIONE DERIVATIVES. Patent number: WO2013071010. Assignee: Allergan, Inc.. Priority date: 11/11/2011. Publication date: 16/05/2013.
32. Escoubet B, Couffignal C, Laisy JP, Mangin L, Chillon S, Laouénan C, Serfaty JM, Jeunemaitre X, Mentré F, Zennaro MC. (2013) Cardiovascular effects of aldosterone: insight from adult carriers of mineralocorticoid receptor mutations. Circ Cardiovasc Genet, 6 (4): 381-90. [PMID:23852419]
33. Fakitsas P, Adam G, Daidié D, van Bemmelen MX, Fouladkou F, Patrignani A, Wagner U, Warth R, Camargo SM, Staub O et al.. (2007) Early aldosterone-induced gene product regulates the epithelial sodium channel by deubiquitylation. J Am Soc Nephrol, 18 (4): 1084-92. [PMID:17344426]
34. Faresse N, Ruffieux-Daidie D, Salamin M, Gomez-Sanchez CE, Staub O. (2010) Mineralocorticoid receptor degradation is promoted by Hsp90 inhibition and the ubiquitin-protein ligase CHIP. Am J Physiol Renal Physiol, 299 (6): F1462-72. [PMID:20861078]
35. Fraccarollo D, Berger S, Galuppo P, Kneitz S, Hein L, Schütz G, Frantz S, Ertl G, Bauersachs J. (2011) Deletion of cardiomyocyte mineralocorticoid receptor ameliorates adverse remodeling after myocardial infarction. Circulation, 123 (4): 400-8. [PMID:21242479]
36. Fuller PJ, Yao YZ, Jin R, He S, Martín-Fernández B, Young MJ, Smith BJ. (2019) Molecular evolution of the switch for progesterone and spironolactone from mineralocorticoid receptor agonist to antagonist. Proc Natl Acad Sci U S A, 116 (37): 18578-18583. [PMID:31439819]
37. Fuse H, Kitagawa H, Kato S. (2000) Characterization of transactivational property and coactivator mediation of rat mineralocorticoid receptor activation function-1 (AF-1). Mol Endocrinol, 14 (6): 889-99. [PMID:10847590]
38. Galigniana MD, Echeverría PC, Erlejman AG, Piwien-Pilipuk G. (2010) Role of molecular chaperones and TPR-domain proteins in the cytoplasmic transport of steroid receptors and their passage through the nuclear pore. Nucleus, 1 (4): 299-308. [PMID:21113270]
39. Galigniana MD, Erlejman AG, Monte M, Gomez-Sanchez C, Piwien-Pilipuk G. (2010) The hsp90-FKBP52 complex links the mineralocorticoid receptor to motor proteins and persists bound to the receptor in early nuclear events. Mol Cell Biol, 30 (5): 1285-98. [PMID:20038533]
40. Gallo LI, Ghini AA, Piwien Pilipuk G, Galigniana MD. (2007) Differential recruitment of tetratricorpeptide repeat domain immunophilins to the mineralocorticoid receptor influences both heat-shock protein 90-dependent retrotransport and hormone-dependent transcriptional activity. Biochemistry, 46 (49): 14044-57. [PMID:18001136]
41. Gasparini S, Resch JM, Narayan SV, Peltekian L, Iverson GN, Karthik S, Geerling JC. (2019) Aldosterone-sensitive HSD2 neurons in mice. Brain Struct Funct, 224 (1): 387-417. [PMID:30343334]
42. Ge RS, Dong Q, Sottas CM, Latif SA, Morris DJ, Hardy MP. (2005) Stimulation of testosterone production in rat Leydig cells by aldosterone is mineralocorticoid receptor mediated. Mol Cell Endocrinol, 243 (1-2): 35-42. [PMID:16188378]
43. Granberg KL, Yuan ZQ, Lindmark B, Edman K, Kajanus J, Hogner A, Malmgren M, O'Mahony G, Nordqvist A, Lindberg J et al.. (2019) Identification of Mineralocorticoid Receptor Modulators with Low Impact on Electrolyte Homeostasis but Maintained Organ Protection. J Med Chem, 62 (3): 1385-1406. [PMID:30596500]
44. Grossmann C, Gekle M. (2012) Interaction between mineralocorticoid receptor and epidermal growth factor receptor signaling. Mol Cell Endocrinol, 350 (2): 235-41. [PMID:21827828]
45. Gumz ML, Popp MP, Wingo CS, Cain BD. (2003) Early transcriptional effects of aldosterone in a mouse inner medullary collecting duct cell line. Am J Physiol Renal Physiol, 285 (4): F664-73. [PMID:12770840]
46. Hellal-Levy C, Couette B, Fagart J, Souque A, Gomez-Sanchez C, Rafestin-Oblin M. (1999) Specific hydroxylations determine selective corticosteroid recognition by human glucocorticoid and mineralocorticoid receptors. FEBS Lett, 464 (1-2): 9-13. [PMID:10611474]
47. Hemmerling M, Nilsson S, Edman K, Eirefelt S, Russell W, Hendrickx R, Johnsson E, Kärrman Mårdh C, Berger M, Rehwinkel H et al.. (2017) Selective Nonsteroidal Glucocorticoid Receptor Modulators for the Inhaled Treatment of Pulmonary Diseases. J Med Chem, 60 (20): 8591-8605. [PMID:28937774]
48. Hong H, Kohli K, Garabedian MJ, Stallcup MR. (1997) GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol, 17 (5): 2735-44. [PMID:9111344]
49. Hudson WH, Youn C, Ortlund EA. (2014) Crystal structure of the mineralocorticoid receptor DNA binding domain in complex with DNA. PLoS ONE, 9 (9): e107000. [PMID:25188500]
50. Hultman ML, Krasnoperova NV, Li S, Du S, Xia C, Dietz JD, Lala DS, Welsch DJ, Hu X. (2005) The ligand-dependent interaction of mineralocorticoid receptor with coactivator and corepressor peptides suggests multiple activation mechanisms. Mol Endocrinol, 19 (6): 1460-73. [PMID:15761029]
51. Iijima T, Katoh M, Takedomi K, Yamamoto Y, Akatsuka H, Shirata N, Nishi A, Takakuwa M, Watanabe Y, Munakata H et al.. (2022) Discovery of Apararenone (MT-3995) as a Highly Selective, Potent, and Novel Nonsteroidal Mineralocorticoid Receptor Antagonist. J Med Chem, 65 (12): 8127-8143. [PMID:35652647]
52. Jaffe IZ, Tintut Y, Newfell BG, Demer LL, Mendelsohn ME. (2007) Mineralocorticoid receptor activation promotes vascular cell calcification. Arterioscler Thromb Vasc Biol, 27 (4): 799-805. [PMID:17234727]
53. Kato M, Furuie H, Shimizu T, Miyazaki A, Kobayashi F, Ishizuka H. (2018) Single- and multiple-dose escalation study to assess pharmacokinetics, pharmacodynamics and safety of oral esaxerenone in healthy Japanese subjects. Br J Clin Pharmacol, 84 (8): 1821-1829. [PMID:29688582]
54. Kitagawa H, Yanagisawa J, Fuse H, Ogawa S, Yogiashi Y, Okuno A, Nagasawa H, Nakajima T, Matsumoto T, Kato S. (2002) Ligand-selective potentiation of rat mineralocorticoid receptor activation function 1 by a CBP-containing histone acetyltransferase complex. Mol Cell Biol, 22 (11): 3698-706. [PMID:11997506]
55. Knutti D, Kaul A, Kralli A. (2000) A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Mol Cell Biol, 20 (7): 2411-22. [PMID:10713165]
56. Kolla V, Litwack G. (2000) Transcriptional regulation of the human Na/K ATPase via the human mineralocorticoid receptor. Mol Cell Biochem, 204 (1-2): 35-40. [PMID:10718622]
57. Kolla V, Robertson NM, Litwack G. (1999) Identification of a mineralocorticoid/glucocorticoid response element in the human Na/K ATPase alpha1 gene promoter. Biochem Biophys Res Commun, 266 (1): 5-14. [PMID:10581156]
58. Krozowski ZS, Funder JW. (1983) Renal mineralocorticoid receptors and hippocampal corticosterone-binding species have identical intrinsic steroid specificity. Proc Natl Acad Sci USA, 80 (19): 6056-60. [PMID:6310613]
59. Kuppusamy M, Gomez-Sanchez EP, Beloate LN, Plonczynski M, Naray-Fejes-Toth A, Fejes-Toth G, Gomez-Sanchez CE. (2017) Interaction of the Mineralocorticoid Receptor With RACK1 and Its Role in Aldosterone Signaling. Endocrinology, 158 (7): 2367-2375. [PMID:28472300]
60. Langer K, Moser D, Otto T, Wolf OT, Kumsta R. (2019) Cortisol modulates the engagement of multiple memory systems: Exploration of a common NR3C2 polymorphism. Psychoneuroendocrinology, 107: 133-140. [PMID:31128569]
61. Le Billan F, Khan JA, Lamribet K, Viengchareun S, Bouligand J, Fagart J, Lombès M. (2015) Cistrome of the aldosterone-activated mineralocorticoid receptor in human renal cells. FASEB J, 29 (9): 3977-89. [PMID:26054365]
62. Lesouhaitier O, Chiappe A, Rossier MF. (2001) Aldosterone increases T-type calcium currents in human adrenocarcinoma (H295R) cells by inducing channel expression. Endocrinology, 142 (10): 4320-30. [PMID:11564691]
63. Liu W, Wang J, Sauter NK, Pearce D. (1995) Steroid receptor heterodimerization demonstrated in vitro and in vivo. Proc Natl Acad Sci USA, 92 (26): 12480-4. [PMID:8618925]
64. McCurley A, Pires PW, Bender SB, Aronovitz M, Zhao MJ, Metzger D, Chambon P, Hill MA, Dorrance AM, Mendelsohn ME et al.. (2012) Direct regulation of blood pressure by smooth muscle cell mineralocorticoid receptors. Nat Med, 18 (9): 1429-33. [PMID:22922412]
65. Murai-Takeda A, Shibata H, Kurihara I, Kobayashi S, Yokota K, Suda N, Mitsuishi Y, Jo R, Kitagawa H, Kato S et al.. (2010) NF-YC functions as a corepressor of agonist-bound mineralocorticoid receptor. J Biol Chem, 285 (11): 8084-93. [PMID:20054001]
66. Nishi M. (2010) Imaging of transcription factor trafficking in living cells: lessons from corticosteroid receptor dynamics. Methods Mol Biol, 647: 199-212. [PMID:20694669]
67. Nishi M, Kawata M. (2007) Dynamics of glucocorticoid receptor and mineralocorticoid receptor: implications from live cell imaging studies. Neuroendocrinology, 85 (3): 186-92. [PMID:17446698]
68. Nishi M, Tanaka M, Matsuda K, Sunaguchi M, Kawata M. (2004) Visualization of glucocorticoid receptor and mineralocorticoid receptor interactions in living cells with GFP-based fluorescence resonance energy transfer. J Neurosci, 24 (21): 4918-27. [PMID:15163683]
69. No authors listed. (2014) Retraction for Kitagawa et al., Ligand-selective potentiation of rat mineralocorticoid receptor activation function 1 by a CBP-containing histone acetyltransferase complex. Mol Cell Biol, 34 (5): 916. [PMID:24509261]
70. Náray-Fejes-Tóth A, Snyder PM, Fejes-Tóth G. (2004) The kidney-specific WNK1 isoform is induced by aldosterone and stimulates epithelial sodium channel-mediated Na+ transport. Proc Natl Acad Sci USA, 101 (50): 17434-9. [PMID:15583131]
71. Oakley RH, Cruz-Topete D, He B, Foley JF, Myers PH, Xu X, Gomez-Sanchez CE, Chambon P, Willis MS, Cidlowski JA. (2019) Cardiomyocyte glucocorticoid and mineralocorticoid receptors directly and antagonistically regulate heart disease in mice. Sci Signal, 12 (577). [PMID:30992401]
72. Obradović D, Tirard M, Némethy Z, Hirsch O, Gronemeyer H, Almeida OF. (2004) DAXX, FLASH, and FAF-1 modulate mineralocorticoid and glucocorticoid receptor-mediated transcription in hippocampal cells--toward a basis for the opposite actions elicited by two nuclear receptors?. Mol Pharmacol, 65 (3): 761-9. [PMID:14978255]
73. Odermatt A, Kratschmar DV. (2012) Tissue-specific modulation of mineralocorticoid receptor function by 11β-hydroxysteroid dehydrogenases: an overview. Mol Cell Endocrinol, 350 (2): 168-86. [PMID:21820034]
74. Ong GSY, Cole TJ, Tesch GH, Morgan J, Dowling JK, Mansell A, Fuller PJ, Young MJ. (2020) Novel mineralocorticoid receptor mechanisms regulate cardiac tissue inflammation in male mice. J Endocrinol, 246 (2): 123-134. [PMID:32464598]
75. Ou XM, Storring JM, Kushwaha N, Albert PR. (2001) Heterodimerization of mineralocorticoid and glucocorticoid receptors at a novel negative response element of the 5-HT1A receptor gene. J Biol Chem, 276 (17): 14299-307. [PMID:11278286]
76. Oñate SA, Tsai SY, Tsai MJ, O'Malley BW. (1995) Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science, 270 (5240): 1354-7. [PMID:7481822]
77. Pascual-Le Tallec L, Demange C, Lombès M. (2004) Human mineralocorticoid receptor A and B protein forms produced by alternative translation sites display different transcriptional activities. Eur J Endocrinol, 150 (4): 585-90. [PMID:15080790]
78. Pascual-Le Tallec L, Simone F, Viengchareun S, Meduri G, Thirman MJ, Lombès M. (2005) The elongation factor ELL (eleven-nineteen lysine-rich leukemia) is a selective coregulator for steroid receptor functions. Mol Endocrinol, 19 (5): 1158-69. [PMID:15650021]
79. Patel PD, Sherman TG, Goldman DJ, Watson SJ. (1989) Molecular cloning of a mineralocorticoid (type I) receptor complementary DNA from rat hippocampus. Mol Endocrinol, 3 (11): 1877-85. [PMID:2558305]
80. Petrovich E, Asher C, Garty H. (2014) Induction of FKBP51 by aldosterone in intestinal epithelium. J Steroid Biochem Mol Biol, 139: 78-87. [PMID:24139875]
81. Piotrowski DW. (2012) Mineralocorticoid receptor antagonists for the treatment of hypertension and diabetic nephropathy. J Med Chem, 55 (18): 7957-66. [PMID:22866979]
82. Pollow K, Juchem M, Elger W, Jacobi N, Hoffmann G, Möbus V. (1992) Dihydrospirorenone (ZK30595): a novel synthetic progestagen--characterization of binding to different receptor proteins. Contraception, 46 (6): 561-74. [PMID:1493716]
83. Poulsen SB, Limbutara K, Fenton RA, Pisitkun T, Christensen BM. (2018) RNA sequencing of kidney distal tubule cells reveals multiple mediators of chronic aldosterone action. Physiol Genomics, 50 (5): 343-354. [PMID:29521601]
84. Pratt WB, Galigniana MD, Morishima Y, Murphy PJ. (2004) Role of molecular chaperones in steroid receptor action. Essays Biochem, 40: 41-58. [PMID:15242338]
85. Pratt WB, Toft DO. (1997) Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev, 18 (3): 306-60. [PMID:9183567]
86. Rafestin-Oblin ME, Couette B, Radanyi C, Lombes M, Baulieu EE. (1989) Mineralocorticosteroid receptor of the chick intestine. Oligomeric structure and transformation. J Biol Chem, 264 (16): 9304-9. [PMID:2542305]
87. Rashmi P, Colussi G, Ng M, Wu X, Kidwai A, Pearce D. (2017) Glucocorticoid-induced leucine zipper protein regulates sodium and potassium balance in the distal nephron. Kidney Int, 91 (5): 1159-1177. [PMID:28094030]
88. Resch JM, Fenselau H, Madara JC, Wu C, Campbell JN, Lyubetskaya A, Dawes BA, Tsai LT, Li MM, Livneh Y et al.. (2017) Aldosterone-Sensing Neurons in the NTS Exhibit State-Dependent Pacemaker Activity and Drive Sodium Appetite via Synergy with Angiotensin II Signaling. Neuron, 96 (1): 190-206.e7. [PMID:28957668]
89. Rickard AJ, Morgan J, Bienvenu LA, Fletcher EK, Cranston GA, Shen JZ, Reichelt ME, Delbridge LM, Young MJ. (2012) Cardiomyocyte mineralocorticoid receptors are essential for deoxycorticosterone/salt-mediated inflammation and cardiac fibrosis. Hypertension, 60 (6): 1443-50. [PMID:23108646]
90. Rickard AJ, Morgan J, Chrissobolis S, Miller AA, Sobey CG, Young MJ. (2014) Endothelial cell mineralocorticoid receptors regulate deoxycorticosterone/salt-mediated cardiac remodeling and vascular reactivity but not blood pressure. Hypertension, 63 (5): 1033-40. [PMID:24566081]
91. Ripa L, Edman K, Dearman M, Edenro G, Hendrickx R, Ullah V, Chang HF, Lepistö M, Chapman D, Geschwindner S et al.. (2018) Discovery of a Novel Oral Glucocorticoid Receptor Modulator (AZD9567) with Improved Side Effect Profile. J Med Chem, 61 (5): 1785-1799. [PMID:29424542]
92. Robert-Nicoud M, Flahaut M, Elalouf JM, Nicod M, Salinas M, Bens M, Doucet A, Wincker P, Artiguenave F, Horisberger JD et al.. (2001) Transcriptome of a mouse kidney cortical collecting duct cell line: effects of aldosterone and vasopressin. Proc Natl Acad Sci U S A, 98 (5): 2712-6. [PMID:11226305]
93. Rogerson FM, Yao YZ, Young MJ, Fuller PJ. (2014) Identification and characterization of a ligand-selective mineralocorticoid receptor coactivator. FASEB J, 28 (10): 4200-10. [PMID:24907116]
94. Ronzaud C, Loffing J, Bleich M, Gretz N, Gröne HJ, Schütz G, Berger S. (2007) Impairment of sodium balance in mice deficient in renal principal cell mineralocorticoid receptor. J Am Soc Nephrol, 18 (6): 1679-87. [PMID:17475815]
95. Rupprecht R, Reul JM, van Steensel B, Spengler D, Söder M, Berning B, Holsboer F, Damm K. (1993) Pharmacological and functional characterization of human mineralocorticoid and glucocorticoid receptor ligands. Eur J Pharmacol, 247 (2): 145-54. [PMID:8282004]
96. Ruzzo EK, Pérez-Cano L, Jung JY, Wang LK, Kashef-Haghighi D, Hartl C, Singh C, Xu J, Hoekstra JN, Leventhal O et al.. (2019) Inherited and De Novo Genetic Risk for Autism Impacts Shared Networks. Cell, 178 (4): 850-866.e26. [PMID:31398340]
97. Sartorato P, Cluzeaud F, Fagart J, Viengchareun S, Lombès M, Zennaro MC. (2004) New naturally occurring missense mutations of the human mineralocorticoid receptor disclose important residues involved in dynamic interactions with deoxyribonucleic acid, intracellular trafficking, and ligand binding. Mol Endocrinol, 18 (9): 2151-65. [PMID:15192075]
98. Schäfer N, Lohmann C, Winnik S, van Tits LJ, Miranda MX, Vergopoulos A, Ruschitzka F, Nussberger J, Berger S, Lüscher TF et al.. (2013) Endothelial mineralocorticoid receptor activation mediates endothelial dysfunction in diet-induced obesity. Eur Heart J, 34 (45): 3515-24. [PMID:23594590]
99. Shen JZ, Morgan J, Tesch GH, Rickard AJ, Chrissobolis S, Drummond GR, Fuller PJ, Young MJ. (2016) Cardiac Tissue Injury and Remodeling Is Dependent Upon MR Regulation of Activation Pathways in Cardiac Tissue Macrophages. Endocrinology, 157 (8): 3213-23. [PMID:27253999]
100. Shibata S, Nagase M, Yoshida S, Kawarazaki W, Kurihara H, Tanaka H, Miyoshi J, Takai Y, Fujita T. (2008) Modification of mineralocorticoid receptor function by Rac1 GTPase: implication in proteinuric kidney disease. Nat Med, 14 (12): 1370-6. [PMID:19029984]
101. Shigaev A, Asher C, Latter H, Garty H, Reuveny E. (2000) Regulation of sgk by aldosterone and its effects on the epithelial Na(+) channel. Am J Physiol Renal Physiol, 278 (4): F613-9. [PMID:10751222]
102. Skrabal F, Auböck J, Edwards CR, Braunsteiner H. (1978) Subtraction potential difference: In-vivo assay for mineralocorticoid activity. Lancet, 1 (8059): 298-302. [PMID:75336]
103. Soundararajan R, Pearce D, Ziera T. (2012) The role of the ENaC-regulatory complex in aldosterone-mediated sodium transport. Mol Cell Endocrinol, 350 (2): 242-7. [PMID:22101317]
104. Soundararajan R, Zhang TT, Wang J, Vandewalle A, Pearce D. (2005) A novel role for glucocorticoid-induced leucine zipper protein in epithelial sodium channel-mediated sodium transport. J Biol Chem, 280 (48): 39970-81. [PMID:16216878]
105. Stephenson G, Krozowski Z, Funder JW. (1984) Extravascular CBG-like sites in rat kidney and mineralocorticoid receptor specificity. Am J Physiol, 246 (2 Pt 2): F227-33. [PMID:6320679]
106. Stockand JD, Spier BJ, Worrell RT, Yue G, Al-Baldawi N, Eaton DC. (1999) Regulation of Na(+) reabsorption by the aldosterone-induced small G protein K-Ras2A. J Biol Chem, 274 (50): 35449-54. [PMID:10585415]
107. Swanson EA, Nelson JW, Jeng S, Erspamer KJ, Yang CL, McWeeney S, Ellison DH. (2019) Salt-sensitive transcriptome of isolated kidney distal tubule cells. Physiol Genomics, 51 (4): 125-135. [PMID:30875275]
108. Tajima T, Kitagawa H, Yokoya S, Tachibana K, Adachi M, Nakae J, Suwa S, Katoh S, Fujieda K. (2000) A novel missense mutation of mineralocorticoid receptor gene in one Japanese family with a renal form of pseudohypoaldosteronism type 1. J Clin Endocrinol Metab, 85 (12): 4690-4. [PMID:11134129]
109. Tallec LP, Kirsh O, Lecomte MC, Viengchareun S, Zennaro MC, Dejean A, Lombès M. (2003) Protein inhibitor of activated signal transducer and activator of transcription 1 interacts with the N-terminal domain of mineralocorticoid receptor and represses its transcriptional activity: implication of small ubiquitin-related modifier 1 modification. Mol Endocrinol, 17 (12): 2529-42. [PMID:14500761]
110. Terker AS, Yarbrough B, Ferdaus MZ, Lazelle RA, Erspamer KJ, Meermeier NP, Park HJ, McCormick JA, Yang CL, Ellison DH. (2016) Direct and Indirect Mineralocorticoid Effects Determine Distal Salt Transport. J Am Soc Nephrol, 27 (8): 2436-45. [PMID:26712527]
111. Trapp T, Rupprecht R, Castrén M, Reul JM, Holsboer F. (1994) Heterodimerization between mineralocorticoid and glucocorticoid receptor: a new principle of glucocorticoid action in the CNS. Neuron, 13 (6): 1457-62. [PMID:7993637]
112. Ueda K, Fujiki K, Shirahige K, Gomez-Sanchez CE, Fujita T, Nangaku M, Nagase M. (2014) Genome-wide analysis of murine renal distal convoluted tubular cells for the target genes of mineralocorticoid receptor. Biochem Biophys Res Commun, 445 (1): 132-7. [PMID:24491541]
113. van Leeuwen N, Caprio M, Blaya C, Fumeron F, Sartorato P, Ronconi V, Giacchetti G, Mantero F, Fernandes-Rosa FL, Simian C et al.. (2010) The functional c.-2G>C variant of the mineralocorticoid receptor modulates blood pressure, renin, and aldosterone levels. Hypertension, 56 (5): 995-1002. [PMID:20855654]
114. van Weert LTCM, Buurstede JC, Mahfouz A, Braakhuis PSM, Polman JAE, Sips HCM, Roozendaal B, Balog J, de Kloet ER, Datson NA et al.. (2017) NeuroD Factors Discriminate Mineralocorticoid From Glucocorticoid Receptor DNA Binding in the Male Rat Brain. Endocrinology, 158 (5): 1511-1522. [PMID:28324065]
115. Verrey F. (1999) Early aldosterone action: toward filling the gap between transcription and transport. Am J Physiol, 277 (3 Pt 2): F319-27. [PMID:10484514]
116. Verrijdt G, Haelens A, Schoenmakers E, Rombauts W, Claessens F. (2002) Comparative analysis of the influence of the high-mobility group box 1 protein on DNA binding and transcriptional activation by the androgen, glucocorticoid, progesterone and mineralocorticoid receptors. Biochem J, 361 (Pt 1): 97-103. [PMID:11742533]
117. Walker BR, Andrew R, Escoubet B, Zennaro MC. (2014) Activation of the hypothalamic-pituitary-adrenal axis in adults with mineralocorticoid receptor haploinsufficiency. J Clin Endocrinol Metab, 99 (8): E1586-91. [PMID:24712576]
118. Wang Q, Anzick S, Richter WF, Meltzer P, Simons SS. (2004) Modulation of transcriptional sensitivity of mineralocorticoid and estrogen receptors. J Steroid Biochem Mol Biol, 91 (4-5): 197-210. [PMID:15336697]
119. Wong S, Brennan FE, Young MJ, Fuller PJ, Cole TJ. (2007) A direct effect of aldosterone on endothelin-1 gene expression in vivo. Endocrinology, 148 (4): 1511-7. [PMID:17218419]
120. Yang J, Fuller PJ, Morgan J, Shibata H, Clyne CD, Young MJ. (2015) GEMIN4 functions as a coregulator of the mineralocorticoid receptor. J Mol Endocrinol, 54 (2): 149-60. [PMID:25555524]
121. Yang J, Fuller PJ, Morgan J, Shibata H, McDonnell DP, Clyne CD, Young MJ. (2014) Use of phage display to identify novel mineralocorticoid receptor-interacting proteins. Mol Endocrinol, 28 (9): 1571-84. [PMID:25000480]
122. Yokota K, Shibata H, Kurihara I, Kobayashi S, Suda N, Murai-Takeda A, Saito I, Kitagawa H, Kato S, Saruta T et al.. (2007) Coactivation of the N-terminal transactivation of mineralocorticoid receptor by Ubc9. J Biol Chem, 282 (3): 1998-2010. [PMID:17105732]
123. Yuan J, Jia R, Bao Y. (2007) Aldosterone up-regulates production of plasminogen activator inhibitor-1 by renal mesangial cells. J Biochem Mol Biol, 40 (2): 180-8. [PMID:17394767]
124. Zennaro MC, Fernandes-Rosa F. (2017) 30 YEARS OF THE MINERALOCORTICOID RECEPTOR: Mineralocorticoid receptor mutations. J Endocrinol, 234 (1): T93-T106. [PMID:28348114]
125. Zennaro MC, Souque A, Viengchareun S, Poisson E, Lombès M. (2001) A new human MR splice variant is a ligand-independent transactivator modulating corticosteroid action. Mol Endocrinol, 15 (9): 1586-98. [PMID:11518808]
126. Zhi L, Ringgenberg JD, Edwards JP, Tegley CM, West SJ, Pio B, Motamedi M, Jones TK, Marschke KB, Mais DE et al.. (2003) Development of progesterone receptor antagonists from 1,2-dihydrochromeno[3,4-f]quinoline agonist pharmacophore. Bioorg Med Chem Lett, 13 (12): 2075-8. [PMID:12781198]
127. Zhou MY, Gomez-Sanchez CE, Gomez-Sanchez EP. (2000) An alternatively spliced rat mineralocorticoid receptor mRNA causing truncation of the steroid binding domain. Mol Cell Endocrinol, 159 (1-2): 125-31. [PMID:10687858]
128. Ziera T, Irlbacher H, Fromm A, Latouche C, Krug SM, Fromm M, Jaisser F, Borden SA. (2009) Cnksr3 is a direct mineralocorticoid receptor target gene and plays a key role in the regulation of the epithelial sodium channel. FASEB J, 23 (11): 3936-46. [PMID:19567370]