
Top ▲

GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
Gene and Protein Information  |
||||||
| class A G protein-coupled receptor | ||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | 7 | 466 | 7q33 | CHRM2 | cholinergic receptor muscarinic 2 | 3,10-11,104,161 |
| Mouse | 7 | 466 | 6 B1 | Chrm2 | cholinergic receptor, muscarinic 2, cardiac | 82 |
| Rat | 7 | 466 | 4q22 | Chrm2 | cholinergic receptor, muscarinic 2 | 138 |
Previous and Unofficial Names  |
| M2 muscarinic acetylcholine receptor | AChR M2 | Chrm-2 | cholinergic receptor, muscarinic 2 | cholinergic receptor | cholinergic receptor, muscarinic 2, cardiac |
Database Links  |
|
| Specialist databases | |
| GPCRdb | acm2_human (Hs), acm2_mouse (Mm), acm2_rat (Rn) |
| Other databases | |
| Alphafold | P08172 (Hs), Q9ERZ4 (Mm), P10980 (Rn) |
| ChEMBL Target | CHEMBL211 (Hs), CHEMBL3197 (Mm), CHEMBL309 (Rn) |
| DrugBank Target | P08172 (Hs) |
| Ensembl Gene | ENSG00000181072 (Hs), ENSMUSG00000045613 (Mm), ENSRNOG00000046972 (Rn) |
| Entrez Gene | 1129 (Hs), 243764 (Mm), 81645 (Rn) |
| Human Protein Atlas | ENSG00000181072 (Hs) |
| KEGG Gene | hsa:1129 (Hs), mmu:243764 (Mm), rno:81645 (Rn) |
| OMIM | 118493 (Hs) |
| Pharos | P08172 (Hs) |
| RefSeq Nucleotide | NM_000739 (Hs), NM_203491 (Mm), NM_031016 (Rn) |
| RefSeq Protein | NP_000730 (Hs), NP_987076 (Mm), NP_112278 (Rn) |
| SynPHARM | 1190 (in complex with [3H]QNB) |
| UniProtKB | P08172 (Hs), Q9ERZ4 (Mm), P10980 (Rn) |
| Wikipedia | CHRM2 (Hs) |
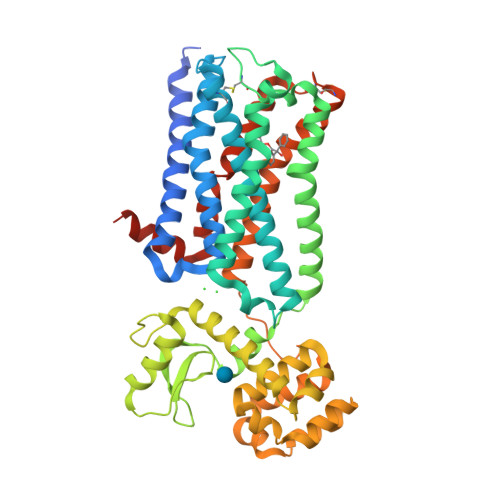
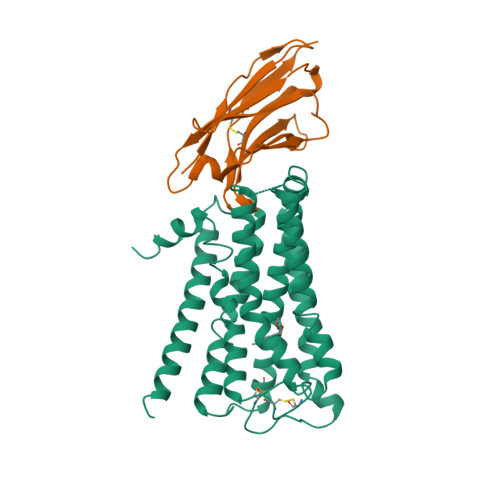
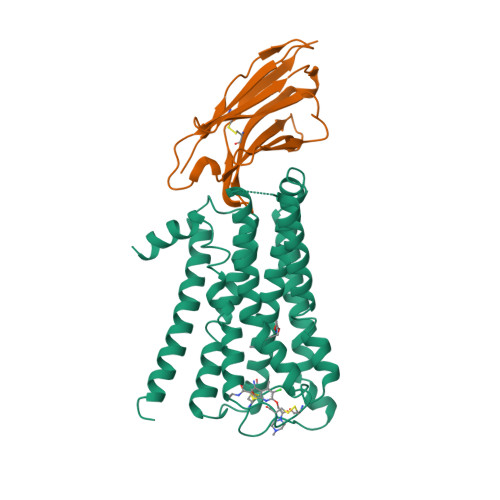
Selected 3D Structures  |
|||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
Download all structure-activity data for this target as a CSV file 
| Agonists | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific agonist tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Agonist Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| The binding data for McN-A-343 [71] is found on rat heart. Please consult references [13,75,110,147,154] for further details of the activity of some of the ligands in this list. Pilocarpine has been found to be a partial agonist [75,154] and a full agonist [147] at the M2 receptor. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antagonists | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific antagonist tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antagonist Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dexetimide is the optical isomer of levetimide [67]. Biperiden is an approved drug antagonist of muscarinic acetylcholine receptors. We have tagged the M1 subtype as the drug's primary target as affinity is 10-fold higher at this receptor subtype [9]. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Allosteric Modulators | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific allosteric modulator tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Primary Transduction Mechanisms 
|
|
| Transducer | Effector/Response |
| Gi/Go family | Adenylyl cyclase inhibition |
| References: 87,103 | |
Secondary Transduction Mechanisms  |
|
| Transducer | Effector/Response |
|
Gs family Gq/G11 family |
Adenylyl cyclase stimulation Phospholipase C stimulation |
| References: 46,86 | |
Tissue Distribution 
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
Expression Datasets  |
|
|
Functional Assays 
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
Physiological Functions 
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
Physiological Consequences of Altering Gene Expression 
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
Phenotypes, Alleles and Disease Models 
|
Mouse data from MGI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Clinically-Relevant Mutations and Pathophysiology 
|
||||||||||
|
||||||||||
| General Comments |
| For reviews on muscarinic receptor knockout mice see [17,84,150-152]. |
1. Anderson DJ, McKinney M. (1988) Muscarinic M2 receptor-mediated cyclic AMP reduction in mechanically dissociated rat cortex. Brain Res, 475 (1): 28-34. [PMID:2850835]
2. Avlani VA, Gregory KJ, Morton CJ, Parker MW, Sexton PM, Christopoulos A. (2007) Critical role for the second extracellular loop in the binding of both orthosteric and allosteric G protein-coupled receptor ligands. J Biol Chem, 282 (35): 25677-86. [PMID:17591774]
3. Badner JA, Yoon SW, Turner G, Bonner TI, Detera-Wadleigh SD. (1995) Multipoint genetic linkage analysis of the m2 human muscarinic receptor gene. Mamm Genome, 6 (7): 489-90. [PMID:7579899]
4. Baghdoyan HA. (1997) Location and quantification of muscarinic receptor subtypes in rat pons: implications for REM sleep generation. Am J Physiol, 273 (3 Pt 2): R896-904. [PMID:9321865]
5. Bernardini N, Roza C, Sauer SK, Gomeza J, Wess J, Reeh PW. (2002) Muscarinic M2 receptors on peripheral nerve endings: a molecular target of antinociception. J Neurosci, 22 (12): RC229. [PMID:12045234]
6. Berrie CP, Birdsall NJ, Hulme EC, Keen M, Stockton JM. (1984) Solubilization and characterization of guanine nucleotide-sensitive muscarinic agonist binding sites from rat myocardium. Br J Pharmacol, 82 (4): 853-61. [PMID:6478115]
7. Birdsall NJ, Farries T, Gharagozloo P, Kobayashi S, Lazareno S, Sugimoto M. (1999) Subtype-selective positive cooperative interactions between brucine analogs and acetylcholine at muscarinic receptors: functional studies. Mol Pharmacol, 55 (4): 778-86. [PMID:10101037]
8. Bognar IT, Beinhauer B, Kann P, Fuder H. (1990) Different muscarinic receptors mediate autoinhibition of acetylcholine release and vagally-induced vasoconstriction in the rat isolated perfused heart. Naunyn Schmiedebergs Arch Pharmacol, 341 (4): 279-87. [PMID:2333099]
9. Bolden C, Cusack B, Richelson E. (1992) Antagonism by antimuscarinic and neuroleptic compounds at the five cloned human muscarinic cholinergic receptors expressed in Chinese hamster ovary cells. J Pharmacol Exp Ther, 260 (2): 576-80. [PMID:1346637]
10. Bonner TI, Modi WS, Seuanez HN, O'Brien SJ. (1991) Chromosomal mapping of the five human genes encoding muscarinic acetylcholine receptors. Cytogenet Cell Genet, 58: 1850-1851.
11. Bonner TI, Young AC, Brann MR, Buckley NJ. (1988) Cloning and expression of the human and rat m5 muscarinic acetylcholine receptor genes. Neuron, 1 (5): 403-10. [PMID:3272174]
12. Brudzynski SM, McLachlan RS, Girvin JP. (1991) Involvement of M1 and M2 muscarinic receptors of the basal forebrain in cholinergically mediated changes in the rat locomotion. Prog Neuropsychopharmacol Biol Psychiatry, 15 (2): 279-84. [PMID:1871329]
13. Bräuner-Osborne H, Ebert B, Brann MR, Falch E, Krogsgaard-Larsen P. (1996) Functional partial agonism at cloned human muscarinic acetylcholine receptors. Eur J Pharmacol, 313 (1-2): 145-50. [PMID:8905341]
14. Buckley NJ, Bonner TI, Buckley CM, Brann MR. (1989) Antagonist binding properties of five cloned muscarinic receptors expressed in CHO-K1 cells. Mol Pharmacol, 35 (4): 469-76. [PMID:2704370]
15. Burke RE. (1986) The relative selectivity of anticholinergic drugs for the M1 and M2 muscarinic receptor subtypes. Mov Disord, 1 (2): 135-44. [PMID:2904117]
16. Bymaster FP, Carter PA, Zhang L, Falcone JF, Stengel PW, Cohen ML, Shannon HE, Gomeza J, Wess J, Felder CC. (2001) Investigations into the physiological role of muscarinic M2 and M4 muscarinic and M4 receptor subtypes using receptor knockout mice. Life Sci, 68 (22-23): 2473-9. [PMID:11392615]
17. Bymaster FP, McKinzie DL, Felder CC, Wess J. (2003) Use of M1-M5 muscarinic receptor knockout mice as novel tools to delineate the physiological roles of the muscarinic cholinergic system. Neurochem Res, 28 (3-4): 437-42. [PMID:12675128]
18. Böhme TM, Keim C, Kreutzmann K, Linder M, Dingermann T, Dannhardt G, Mutschler E, Lambrecht G. (2003) Structure-activity relationships of dimethindene derivatives as new M2-selective muscarinic receptor antagonists. J Med Chem, 46 (5): 856-67. [PMID:12593665]
19. Candell LM, Yun SH, Tran LL, Ehlert FJ. (1990) Differential coupling of subtypes of the muscarinic receptor to adenylate cyclase and phosphoinositide hydrolysis in the longitudinal muscle of the rat ileum. Mol Pharmacol, 38 (5): 689-97. [PMID:2172776]
20. Cembala TM, Sherwin JD, Tidmarsh MD, Appadu BL, Lambert DG. (1998) Interaction of neuromuscular blocking drugs with recombinant human m1-m5 muscarinic receptors expressed in Chinese hamster ovary cells. Br J Pharmacol, 125 (5): 1088-94. [PMID:9846649]
21. Ch'ng SS, Walker AJ, McCarthy M, Le TK, Thomas N, Gibbons A, Udawela M, Kusljic S, Dean B, Gogos A. (2020) The Impact of Removal of Ovarian Hormones on Cholinergic Muscarinic Receptors: Examining Prepulse Inhibition and Receptor Binding. Brain Sci, 10 (2). [PMID:32079174]
22. Chariot J, De la Tour J, Vaille C, Rozé C. (1987) Comparative effects of pirenzepine and atropine on pancreatic secretion in conscious rats. Arch Int Pharmacodyn Ther, 285 (1): 158-65. [PMID:3579423]
23. Cheng K, Khurana S, Chen Y, Kennedy RH, Zimniak P, Raufman JP. (2002) Lithocholylcholine, a bile acid/acetylcholine hybrid, is a muscarinic receptor antagonist. J Pharmacol Exp Ther, 303 (1): 29-35. [PMID:12235229]
24. Christopoulos A, Grant MK, Ayoubzadeh N, Kim ON, Sauerberg P, Jeppesen L, El-Fakahany EE. (2001) Synthesis and pharmacological evaluation of dimeric muscarinic acetylcholine receptor agonists. J Pharmacol Exp Ther, 298 (3): 1260-8. [PMID:11504829]
25. Christopoulos A, Sorman JL, Mitchelson F, El-Fakahany EE. (1999) Characterization of the subtype selectivity of the allosteric modulator heptane-1,7-bis-(dimethyl-3'-phthalimidopropyl) ammonium bromide (C7/3-phth) at cloned muscarinic acetylcholine receptors. Biochem Pharmacol, 57 (2): 171-9. [PMID:9890565]
26. Clark AL, Mitchelson F. (1976) The inhibitory effect of gallamine on muscarinic receptors. Br J Pharmacol, 58 (3): 323-31. [PMID:990587]
27. Cortés R, Probst A, Tobler HJ, Palacios JM. (1986) Muscarinic cholinergic receptor subtypes in the human brain. II. Quantitative autoradiographic studies. Brain Res, 362 (2): 239-53. [PMID:3753655]
28. Croy CH, Chan WY, Castetter AM, Watt ML, Quets AT, Felder CC. (2016) Characterization of PCS1055, a novel muscarinic M4 receptor antagonist. Eur J Pharmacol, 782: 70-6. [PMID:27085897]
29. Croy CH, Schober DA, Xiao H, Quets A, Christopoulos A, Felder CC. (2014) Characterization of the novel positive allosteric modulator, LY2119620, at the muscarinic M(2) and M(4) receptors. Mol Pharmacol, 86 (1): 106-15. [PMID:24807965]
30. Del Bello F, Barocelli E, Bertoni S, Bonifazi A, Camalli M, Campi G, Giannella M, Matucci R, Nesi M, Pigini M et al.. (2012) 1,4-dioxane, a suitable scaffold for the development of novel M₃ muscarinic receptor antagonists. J Med Chem, 55 (4): 1783-7. [PMID:22243489]
31. Disse B, Reichl R, Speck G, Traunecker W, Ludwig Rominger KL, Hammer R. (1993) Ba 679 BR, a novel long-acting anticholinergic bronchodilator. Life Sci, 52 (5-6): 537-44. [PMID:8441333]
32. Duttaroy A, Gomeza J, Gan JW, Siddiqui N, Basile AS, Harman WD, Smith PL, Felder CC, Levey AI, Wess J. (2002) Evaluation of muscarinic agonist-induced analgesia in muscarinic acetylcholine receptor knockout mice. Mol Pharmacol, 62 (5): 1084-93. [PMID:12391271]
33. Dörje F, Wess J, Lambrecht G, Tacke R, Mutschler E, Brann MR. (1991) Antagonist binding profiles of five cloned human muscarinic receptor subtypes. J Pharmacol Exp Ther, 256 (2): 727-33. [PMID:1994002]
34. Ehlert FJ, Griffin MT, Abe DM, Vo TH, Taketo MM, Manabe T, Matsui M. (2005) The M2 muscarinic receptor mediates contraction through indirect mechanisms in mouse urinary bladder. J Pharmacol Exp Ther, 313 (1): 368-78. [PMID:15608083]
35. Ehlert FJ, Griffin MT, Glidden PF. (1996) The interaction of the enantiomers of aceclidine with subtypes of the muscarinic receptor. J Pharmacol Exp Ther, 279 (3): 1335-44. [PMID:8968358]
36. Ehlert FJ, Rathbun BE. (1990) Signaling through the muscarinic receptor-adenylate cyclase system of the heart is buffered against GTP over a range of concentrations. Mol Pharmacol, 38 (1): 148-58. [PMID:2370853]
37. Esqueda EE, Gerstin Jr EH, Griffin MT, Ehlert FJ. (1996) Stimulation of cyclic AMP accumulation and phosphoinositide hydrolysis by M3 muscarinic receptors in the rat peripheral lung. Biochem Pharmacol, 52 (4): 643-58. [PMID:8759038]
38. Fargeas MJ, Fioramonti J, Buéno L. (1987) Central muscarinic control of the pattern of small intestinal motility in rats. Life Sci, 40 (17): 1709-15. [PMID:3561171]
39. Fernandez-Fernandez JM, Wanaverbecq N, Halley P, Caulfield MP, Brown DA. (1999) Selective activation of heterologously expressed G protein-gated K+ channels by M2 muscarinic receptors in rat sympathetic neurones. J Physiol (Lond.), 515 ( Pt 3): 631-7. [PMID:10066893]
40. Fernández-Fernández JM, Abogadie FC, Milligan G, Delmas P, Brown DA. (2001) Multiple pertussis toxin-sensitive G-proteins can couple receptors to GIRK channels in rat sympathetic neurons when expressed heterologously, but only native G(i)-proteins do so in situ. Eur J Neurosci, 14 (2): 283-92. [PMID:11553279]
41. Fisher JT, Vincent SG, Gomeza J, Yamada M, Wess J. (2004) Loss of vagally mediated bradycardia and bronchoconstriction in mice lacking M2 or M3 muscarinic acetylcholine receptors. FASEB J, 18 (6): 711-3. [PMID:14977875]
42. Gentry PR, Kokubo M, Bridges TM, Cho HP, Smith E, Chase P, Hodder PS, Utley TJ, Rajapakse A, Byers F et al.. (2014) Discovery, synthesis and characterization of a highly muscarinic acetylcholine receptor (mAChR)-selective M5-orthosteric antagonist, VU0488130 (ML381): a novel molecular probe. ChemMedChem, 9 (8): 1677-82. [PMID:24692176]
43. Gillberg PG, Gordh Jr T, Hartvig P, Jansson I, Pettersson J, Post C. (1989) Characterization of the antinociception induced by intrathecally administered carbachol. Pharmacol Toxicol, 64 (4): 340-3. [PMID:2748539]
44. Gillberg PG, Sundquist S, Nilvebrant L. (1998) Comparison of the in vitro and in vivo profiles of tolterodine with those of subtype-selective muscarinic receptor antagonists. Eur J Pharmacol, 349 (2-3): 285-92. [PMID:9671109]
45. Gomeza J, Shannon H, Kostenis E, Felder C, Zhang L, Brodkin J, Grinberg A, Sheng H, Wess J. (1999) Pronounced pharmacologic deficits in M2 muscarinic acetylcholine receptor knockout mice. Proc Natl Acad Sci USA, 96 (4): 1692-7. [PMID:9990086]
46. Griffin MT, Figueroa KW, Liller S, Ehlert FJ. (2007) Estimation of agonist activity at G protein-coupled receptors: analysis of M2 muscarinic receptor signaling through Gi/o,Gs, and G15. J Pharmacol Exp Ther, 321 (3): 1193-207. [PMID:17392404]
47. Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T et al.. (2012) Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature, 482 (7386): 547-51. [PMID:22278061]
48. Hagan JJ, Tonnaer JA, Broekkamp CL. (1987) Cholinergic stimulation of drinking from the lateral hypothalamus: indications for M2 muscarinic receptor mediation. Pharmacol Biochem Behav, 26 (4): 771-9. [PMID:3602034]
49. Hassall CJ, Stanford SC, Burnstock G, Buckley NJ. (1993) Co-expression of four muscarinic receptor genes by the intrinsic neurons of the rat and guinea-pig heart. Neuroscience, 56 (4): 1041-8. [PMID:8284034]
50. Hegde SS, Choppin A, Bonhaus D, Briaud S, Loeb M, Moy TM, Loury D, Eglen RM. (1997) Functional role of M2 and M3 muscarinic receptors in the urinary bladder of rats in vitro and in vivo. Br J Pharmacol, 120 (8): 1409-18. [PMID:9113359]
51. Hegde SS, Pulido-Rios MT, Luttmann MA, Foley JJ, Hunsberger GE, Steinfeld T, Lee T, Ji Y, Mammen MM, Jasper JR. (2018) Pharmacological properties of revefenacin (TD-4208), a novel, nebulized long-acting, and lung selective muscarinic antagonist, at human recombinant muscarinic receptors and in rat, guinea pig, and human isolated airway tissues. Pharmacol Res Perspect, 6 (3): e00400. [PMID:29736245]
52. Hirose H, Aoki I, Kimura T, Fujikawa T, Numazawa T, Sasaki K, Sato A, Hasegawa T, Nishikibe M, Mitsuya M et al.. (2001) Pharmacological properties of (2R)-N-[1-(6-aminopyridin-2-ylmethyl)piperidin-4-yl]-2-[(1R)-3,3-difluorocyclopentyl]-2-hydroxy-2-phenylacetamide: a novel mucarinic antagonist with M(2)-sparing antagonistic activity. J Pharmacol Exp Ther, 297 (2): 790-7. [PMID:11303071]
53. Hohmann CF, Potter ED, Levey AI. (1995) Development of muscarinic receptor subtypes in the forebrain of the mouse. J Comp Neurol, 358 (1): 88-101. [PMID:7560279]
54. Huang F, Buchwald P, Browne CE, Farag HH, Wu WM, Ji F, Hochhaus G, Bodor N. (2001) Receptor binding studies of soft anticholinergic agents. AAPS PharmSci, 3 (4): E30. [PMID:12049493]
55. Ikeda K, Kobayashi S, Suzuki M, Miyata K, Takeuchi M, Yamada T, Honda K. (2002) M(3) receptor antagonism by the novel antimuscarinic agent solifenacin in the urinary bladder and salivary gland. Naunyn Schmiedebergs Arch Pharmacol, 366 (2): 97-103. [PMID:12122494]
56. Imeri L, Bianchi S, Angeli P, Mancia M. (1991) Differential effects of M2 and M3 muscarinic antagonists on the sleep-wake cycle. Neuroreport, 2 (7): 383-5. [PMID:1912471]
57. Imeri L, Bianchi S, Angeli P, Mancia M. (1994) Selective blockade of different brain stem muscarinic receptor subtypes: effects on the sleep-wake cycle. Brain Res, 636 (1): 68-72. [PMID:8156412]
58. Imeri L, Bianchi S, Angeli P, Mancia M. (1996) Muscarinic receptor subtypes in the medial preoptic area and sleep-wake cycles. Neuroreport, 7 (2): 417-20. [PMID:8730795]
59. Jagoda EM, Kiesewetter DO, Shimoji K, Ravasi L, Yamada M, Gomeza J, Wess J, Eckelman WC. (2003) Regional brain uptake of the muscarinic ligand, [18F]FP-TZTP, is greatly decreased in M2 receptor knockout mice but not in M1, M3 and M4 receptor knockout mice. Neuropharmacology, 44 (5): 653-61. [PMID:12668051]
60. Jakubík J, Bacáková L, el-Fakahany EE, Tucek S. (1995) Subtype selectivity of the positive allosteric action of alcuronium at cloned M1-M5 muscarinic acetylcholine receptors. J Pharmacol Exp Ther, 274 (3): 1077-83. [PMID:7562472]
61. Jakubík J, Bacáková L, El-Fakahany EE, Tucek S. (1997) Positive cooperativity of acetylcholine and other agonists with allosteric ligands on muscarinic acetylcholine receptors. Mol Pharmacol, 52 (1): 172-9. [PMID:9224827]
62. Jakubík J, El-Fakahany EE, Dolezal V. (2006) Differences in kinetics of xanomeline binding and selectivity of activation of G proteins at M(1) and M(2) muscarinic acetylcholine receptors. Mol Pharmacol, 70 (2): 656-66. [PMID:16675658]
63. Jolkkonen M, van Giersbergen PL, Hellman U, Wernstedt C, Karlsson E. (1994) A toxin from the green mamba Dendroaspis angusticeps: amino acid sequence and selectivity for muscarinic m4 receptors. FEBS Lett, 352 (1): 91-4. [PMID:7925952]
64. Kashihara K, Varga EV, Waite SL, Roeske WR, Yamamura HI. (1992) Cloning of the rat M3, M4 and M5 muscarinic acetylcholine receptor genes by the polymerase chain reaction (PCR) and the pharmacological characterization of the expressed genes. Life Sci, 51 (12): 955-71. [PMID:1325587]
65. Keov P, Valant C, Devine SM, Lane JR, Scammells PJ, Sexton PM, Christopoulos A. (2013) Reverse engineering of the selective agonist TBPB unveils both orthosteric and allosteric modes of action at the M₁ muscarinic acetylcholine receptor. Mol Pharmacol, 84 (3): 425-37. [PMID:23798605]
66. Khattar SK, Bora RS, Priyadarsiny P, Gupta D, Khanna A, Narayanan KL, Babu V, Chugh A, Saini KS. (2006) High level stable expression of pharmacologically active human M1-M5 muscarinic receptor subtypes in mammalian cells. Biotechnol Lett, 28 (2): 121-9. [PMID:16369696]
67. Kovacs I, Yamamura HI, Waite SL, Varga EV, Roeske WR. (1998) Pharmacological comparison of the cloned human and rat M2 muscarinic receptor genes expressed in the murine fibroblast (B82) cell line. J Pharmacol Exp Ther, 284 (2): 500-7. [PMID:9454790]
68. Kruse AC, Ring AM, Manglik A, Hu J, Hu K, Eitel K, Hübner H, Pardon E, Valant C, Sexton PM et al.. (2013) Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature, 504 (7478): 101-6. [PMID:24256733]
69. Lachowicz JE, Lowe D, Duffy RA, Ruperto V, Taylor LA, Guzik H, Brown J, Berger JG, Tice M, McQuade R et al.. (1999) SCH 57790: a novel M2 receptor selective antagonist. Life Sci, 64 (6-7): 535-9. [PMID:10069520]
70. Lainé DI, McCleland B, Thomas S, Neipp C, Underwood B, Dufour J, Widdowson KL, Palovich MR, Blaney FE, Foley JJ et al.. (2009) Discovery of novel 1-azoniabicyclo[2.2.2]octane muscarinic acetylcholine receptor antagonists. J Med Chem, 52 (8): 2493-505. [PMID:19317446]
71. Lambrecht G, Moser U, Grimm U, Pfaff O, Hermanni U, Hildebrandt C, Waelbroeck M, Christophe J, Mutschler E. (1993) New functionally selective muscarinic agonists. Life Sci, 52 (5-6): 481-8. [PMID:7680092]
72. Lazareno S, Birdsall NJ. (1993) Pharmacological characterization of acetylcholine-stimulated [35S]-GTP gamma S binding mediated by human muscarinic m1-m4 receptors: antagonist studies. Br J Pharmacol, 109 (4): 1120-7. [PMID:8401923]
73. Lazareno S, Birdsall NJ. (1995) Detection, quantitation, and verification of allosteric interactions of agents with labeled and unlabeled ligands at G protein-coupled receptors: interactions of strychnine and acetylcholine at muscarinic receptors. Mol Pharmacol, 48 (2): 362-78. [PMID:7651370]
74. Lazareno S, Dolezal V, Popham A, Birdsall NJ. (2004) Thiochrome enhances acetylcholine affinity at muscarinic M4 receptors: receptor subtype selectivity via cooperativity rather than affinity. Mol Pharmacol, 65 (1): 257-66. [PMID:14722259]
75. Lazareno S, Farries T, Birdsall NJ. (1993) Pharmacological characterization of guanine nucleotide exchange reactions in membranes from CHO cells stably transfected with human muscarinic receptors m1-m4. Life Sci, 52 (5-6): 449-56. [PMID:8441327]
76. Lazareno S, Gharagozloo P, Kuonen D, Popham A, Birdsall NJ. (1998) Subtype-selective positive cooperative interactions between brucine analogues and acetylcholine at muscarinic receptors: radioligand binding studies. Mol Pharmacol, 53 (3): 573-89. [PMID:9495826]
77. Lazareno S, Popham A, Birdsall NJ. (2000) Allosteric interactions of staurosporine and other indolocarbazoles with N-[methyl-(3)H]scopolamine and acetylcholine at muscarinic receptor subtypes: identification of a second allosteric site. Mol Pharmacol, 58 (1): 194-207. [PMID:10860942]
78. Lazareno S, Popham A, Birdsall NJ. (2002) Analogs of WIN 62,577 define a second allosteric site on muscarinic receptors. Mol Pharmacol, 62 (6): 1492-505. [PMID:12435818]
79. Levey AI, Edmunds SM, Koliatsos V, Wiley RG, Heilman CJ. (1995) Expression of m1-m4 muscarinic acetylcholine receptor proteins in rat hippocampus and regulation by cholinergic innervation. J Neurosci, 15 (5 Pt 2): 4077-92. [PMID:7751967]
80. Loudon JM, Bromidge SM, Brown F, Clark MS, Hatcher JP, Hawkins J, Riley GJ, Noy G, Orlek BS. (1997) SB 202026: a novel muscarinic partial agonist with functional selectivity for M1 receptors. J Pharmacol Exp Ther, 283 (3): 1059-68. [PMID:9399977]
81. Maggio R, Barbier P, Bolognesi ML, Minarini A, Tedeschi D, Melchiorre C. (1994) Binding profile of the selective muscarinic receptor antagonist tripitramine. Eur J Pharmacol, 268: 459-462. [PMID:7805774]
82. Matsui M, Araki Y, Karasawa H, Matsubara N, Taketo MM, Seldin MF. (1999) Mapping of five subtype genes for muscarinic acetylcholine receptor to mouse chromosomes. Genes Genet Syst, 74 (1): 15-21. [PMID:10549128]
83. Matsui M, Griffin MT, Shehnaz D, Taketo MM, Ehlert FJ. (2003) Increased relaxant action of forskolin and isoproterenol against muscarinic agonist-induced contractions in smooth muscle from M2 receptor knockout mice. J Pharmacol Exp Ther, 305 (1): 106-13. [PMID:12649358]
84. Matsui M, Yamada S, Oki T, Manabe T, Taketo MM, Ehlert FJ. (2004) Functional analysis of muscarinic acetylcholine receptors using knockout mice. Life Sci, 75 (25): 2971-81. [PMID:15474550]
85. McCormack DG, Mak JC, Minette P, Barnes PJ. (1988) Muscarinic receptor subtypes mediating vasodilation in the pulmonary artery. Eur J Pharmacol, 158 (3): 293-7. [PMID:3253104]
86. Michal P, El-Fakahany EE, Dolezal V. (2007) Muscarinic M2 receptors directly activate Gq/11 and Gs G-proteins. J Pharmacol Exp Ther, 320 (2): 607-14. [PMID:17065363]
87. Migeon JC, Thomas SL, Nathanson NM. (1995) Differential coupling of m2 and m4 muscarinic receptors to inhibition of adenylyl cyclase by Gi alpha and G(o)alpha subunits. J Biol Chem, 270 (27): 16070-4. [PMID:7608168]
88. Miller JH, Aagaard PJ, Gibson VA, McKinney M. (1992) Binding and functional selectivity of himbacine for cloned and neuronal muscarinic receptors. J Pharmacol Exp Ther, 263 (2): 663-7. [PMID:1331410]
89. Miller JH, Gibson VA, McKinney M. (1991) Binding of [3H]AF-DX 384 to cloned and native muscarinic receptors. J Pharmacol Exp Ther, 259 (2): 601-7. [PMID:1941609]
90. Minette PA, Barnes PJ. (1988) Prejunctional inhibitory muscarinic receptors on cholinergic nerves in human and guinea pig airways. J Appl Physiol, 64 (6): 2532-7. [PMID:3403437]
91. Mohr M, Heller E, Ataie A, Mohr K, Holzgrabe U. (2004) Development of a new type of allosteric modulator of muscarinic receptors: hybrids of the antagonist AF-DX 384 and the hexamethonio derivative W84. J Med Chem, 47 (12): 3324-7. [PMID:15163212]
92. Moo EV, Sexton PM, Christopoulos A, Valant C. (2018) Utility of an "Allosteric Site-Impaired" M2 Muscarinic Acetylcholine Receptor as a Novel Construct for Validating Mechanisms of Action of Synthetic and Putative Endogenous Allosteric Modulators. Mol Pharmacol, 94 (5): 1298-1309. [PMID:30213802]
93. Narducci F, Bassotti G, Daniotti S, Del Soldato P, Pelli MA, Morelli A. (1985) Identification of muscarinic receptor subtype mediating colonic response to eating. Dig Dis Sci, 30 (2): 124-8. [PMID:3838152]
94. Nelson CP, Nahorski SR, Challiss RA. (2006) Constitutive activity and inverse agonism at the M2 muscarinic acetylcholine receptor. J Pharmacol Exp Ther, 316 (1): 279-88. [PMID:16188951]
95. Nenasheva TA, Neary M, Mashanov GI, Birdsall NJ, Breckenridge RA, Molloy JE. (2013) Abundance, distribution, mobility and oligomeric state of M₂ muscarinic acetylcholine receptors in live cardiac muscle. J Mol Cell Cardiol, 57: 129-36. [PMID:23357106]
96. Newman AH, Kline RH, Allen AC, Izenwasser S, George C, Katz JL. (1995) Novel 4'-substituted and 4',4"-disubstituted 3 alpha-(diphenylmethoxy)tropane analogs as potent and selective dopamine uptake inhibitors. J Med Chem, 38 (20): 3933-40. [PMID:7562926]
97. Näsman J, Jolkkonen M, Ammoun S, Karlsson E, Akerman KE. (2000) Recombinant expression of a selective blocker of M(1) muscarinic receptors. Biochem Biophys Res Commun, 271 (2): 435-9. [PMID:10799315]
98. Oki T, Takagi Y, Inagaki S, Taketo MM, Manabe T, Matsui M, Yamada S. (2005) Quantitative analysis of binding parameters of [3H]N-methylscopolamine in central nervous system of muscarinic acetylcholine receptor knockout mice. Brain Res Mol Brain Res, 133 (1): 6-11. [PMID:15661360]
99. Olianas MC, Ingianni A, Maullu C, Adem A, Karlsson E, Onali P. (1999) Selectivity profile of muscarinic toxin 3 in functional assays of cloned and native receptors. J Pharmacol Exp Ther, 288 (1): 164-70. [PMID:9862767]
100. Ozenil M, Pacher K, Balber T, Vraka C, Roller A, Holzer W, Spreitzer H, Mitterhauser M, Wadsak W, Hacker M et al.. (2020) Enhanced arecoline derivatives as muscarinic acetylcholine receptor M1 ligands for potential application as PET radiotracers. Eur J Med Chem, 204: 112623. [PMID:32717485]
101. Pazos A, Wiederhold KH, Palacios JM. (1986) Central pressor effects induced by muscarinic receptor agonists: evidence for a predominant role of the M2 receptor subtype. Eur J Pharmacol, 125 (1): 63-70. [PMID:3732392]
102. Pei XF, Gupta TH, Badio B, Padgett WL, Daly JW. (1998) 6beta-Acetoxynortropane: a potent muscarinic agonist with apparent selectivity toward M2-receptors. J Med Chem, 41 (12): 2047-55. [PMID:9622546]
103. Peralta EG, Ashkenazi A, Winslow JW, Ramachandran J, Capon DJ. (1988) Differential regulation of PI hydrolysis and adenylyl cyclase by muscarinic receptor subtypes. Nature, 334 (6181): 434-7. [PMID:2841607]
104. Peralta EG, Ashkenazi A, Winslow JW, Smith DH, Ramachandran J, Capon DJ. (1987) Distinct primary structures, ligand-binding properties and tissue-specific expression of four human muscarinic acetylcholine receptors. EMBO J, 6 (13): 3923-9. [PMID:3443095]
105. Powers AS, Pham V, Burger WAC, Thompson G, Laloudakis Y, Barnes NW, Sexton PM, Paul SM, Christopoulos A, Thal DM et al.. (2023) Structural basis of efficacy-driven ligand selectivity at GPCRs. Nat Chem Biol, 19 (7): 805-814. [PMID:36782010]
106. Prat M, Buil MA, Fernández MD, Castro J, Monleón JM, Tort L, Casals G, Ferrer M, Huerta JM, Espinosa S et al.. (2011) Discovery of novel quaternary ammonium derivatives of (3R)-quinuclidinyl carbamates as potent and long acting muscarinic antagonists. Bioorg Med Chem Lett, 21 (11): 3457-61. [PMID:21524581]
107. Prat M, Fernández D, Buil MA, Crespo MI, Casals G, Ferrer M, Tort L, Castro J, Monleón JM, Gavaldà A et al.. (2009) Discovery of novel quaternary ammonium derivatives of (3R)-quinuclidinol esters as potent and long-acting muscarinic antagonists with potential for minimal systemic exposure after inhaled administration: identification of (3R)-3-{[hydroxy(di-2-thienyl)acetyl]oxy}-1-(3-phenoxypropyl)-1-azoniabicyclo[2.2.2]octane bromide (aclidinium bromide). J Med Chem, 52 (16): 5076-92. [PMID:19653626]
108. Preiksaitis HG, Krysiak PS, Chrones T, Rajgopal V, Laurier LG. (2000) Pharmacological and molecular characterization of muscarinic receptor subtypes in human esophageal smooth muscle. J Pharmacol Exp Ther, 295 (3): 879-88. [PMID:11082420]
109. Rhodes ME, Billings TE, Czambel RK, Rubin RT. (2005) Pituitary-adrenal responses to cholinergic stimulation and acute mild stress are differentially elevated in male and female M(2) muscarinic receptor knockout mice. J Neuroendocrinol, 17 (12): 817-26. [PMID:16280029]
110. Richards MH, van Giersbergen PL. (1995) Human muscarinic receptors expressed in A9L and CHO cells: activation by full and partial agonists. Br J Pharmacol, 114 (6): 1241-9. [PMID:7620715]
111. Rosenblum K, Futter M, Jones M, Hulme EC, Bliss TV. (2000) ERKI/II regulation by the muscarinic acetylcholine receptors in neurons. J Neurosci, 20 (3): 977-85. [PMID:10648702]
112. Salmon M, Luttmann MA, Foley JJ, Buckley PT, Schmidt DB, Burman M, Webb EF, DeHaas CJ, Kotzer CJ, Barrett VJ et al.. (2013) Pharmacological characterization of GSK573719 (umeclidinium): a novel, long-acting, inhaled antagonist of the muscarinic cholinergic receptors for treatment of pulmonary diseases. J Pharmacol Exp Ther, 345 (2): 260-70. [PMID:23435542]
113. Sandvik AK, Kleveland PM, Waldum HL. (1988) Muscarinic M2 stimulation releases histamine in the totally isolated, vascularly perfused rat stomach. Scand J Gastroenterol, 23 (9): 1049-56. [PMID:2470130]
114. Schrage R, Holze J, Klöckner J, Balkow A, Klause AS, Schmitz AL, De Amici M, Kostenis E, Tränkle C, Holzgrabe U et al.. (2014) New insight into active muscarinic receptors with the novel radioagonist [³H]iperoxo. Biochem Pharmacol, 90 (3): 307-19. [PMID:24863257]
115. Schrage R, Seemann WK, Klöckner J, Dallanoce C, Racké K, Kostenis E, De Amici M, Holzgrabe U, Mohr K. (2013) Agonists with supraphysiological efficacy at the muscarinic M2 ACh receptor. Br J Pharmacol, 169 (2): 357-70. [PMID:23062057]
116. Seeger T, Fedorova I, Zheng F, Miyakawa T, Koustova E, Gomeza J, Basile AS, Alzheimer C, Wess J. (2004) M2 muscarinic acetylcholine receptor knock-out mice show deficits in behavioral flexibility, working memory, and hippocampal plasticity. J Neurosci, 24 (45): 10117-27. [PMID:15537882]
117. Sen AP, Bhattacharya SK. (1991) Thermic response of selective muscarinic agonists and antagonists in rat. Indian J Exp Biol, 29 (2): 131-5. [PMID:1869296]
118. Shapiro RA, Scherer NM, Habecker BA, Subers EM, Nathanson NM. (1988) Isolation, sequence, and functional expression of the mouse M1 muscarinic acetylcholine receptor gene. J Biol Chem, 263 (34): 18397-403. [PMID:2848036]
119. Sheffler DJ, Williams R, Bridges TM, Xiang Z, Kane AS, Byun NE, Jadhav S, Mock MM, Zheng F, Lewis LM et al.. (2009) A novel selective muscarinic acetylcholine receptor subtype 1 antagonist reduces seizures without impairing hippocampus-dependent learning. Mol Pharmacol, 76 (2): 356-68. [PMID:19407080]
120. Shida T, Tokunaga A, Kondo E, Ueda Y, Ohno K, Saika T, Kiyama H, Tohyama M. (1993) Expression of muscarinic and nicotinic receptor mRNA in the salivary gland of rats: a study by in situ hybridization histochemistry. Brain Res Mol Brain Res, 17 (3-4): 335-9. [PMID:8510505]
121. Sinha S, Gupta S, Malhotra S, Krishna NS, Meru AV, Babu V, Bansal V, Garg M, Kumar N, Chugh A et al.. (2010) AE9C90CB: a novel, bladder-selective muscarinic receptor antagonist for the treatment of overactive bladder. Br J Pharmacol, 160 (5): 1119-27. [PMID:20590605]
122. Sowell Sr JW, Tang Y, Valli MJ, Chapman Jr JM, Usher LA, Vaughan CM, Kosh JW. (1992) Synthesis and cholinergic properties of bis[[(dimethylamino)methyl]furanyl] analogues of ranitidine. J Med Chem, 35 (6): 1102-8. [PMID:1552502]
123. Stanton T, Bolden-Watson C, Cusack B, Richelson E. (1993) Antagonism of the five cloned human muscarinic cholinergic receptors expressed in CHO-K1 cells by antidepressants and antihistaminics. Biochem Pharmacol, 45 (11): 2352-4. [PMID:8100134]
124. Stein R, Pinkas-Kramarski R, Sokolovsky M. (1988) Cloned M1 muscarinic receptors mediate both adenylate cyclase inhibition and phosphoinositide turnover. EMBO J, 7 (10): 3031-5. [PMID:2846274]
125. Steinfeld T, Mammen M, Smith JA, Wilson RD, Jasper JR. (2007) A novel multivalent ligand that bridges the allosteric and orthosteric binding sites of the M2 muscarinic receptor. Mol Pharmacol, 72 (2): 291-302. [PMID:17478612]
126. Stengel PW, Cohen ML. (2002) Muscarinic receptor knockout mice: role of muscarinic acetylcholine receptors M(2), M(3), and M(4) in carbamylcholine-induced gallbladder contractility. J Pharmacol Exp Ther, 301 (2): 643-50. [PMID:11961069]
127. Stengel PW, Gomeza J, Wess J, Cohen ML. (2000) M(2) and M(4) receptor knockout mice: muscarinic receptor function in cardiac and smooth muscle in vitro. J Pharmacol Exp Ther, 292 (3): 877-85. [PMID:10688600]
128. Stocks MJ, Alcaraz L, Bailey A, Bowers K, Donald D, Edwards H, Hunt F, Kindon N, Pairaudeau G, Theaker J et al.. (2010) The discovery of new spirocyclic muscarinic M3 antagonists. Bioorg Med Chem Lett, 20 (24): 7458-61. [PMID:21036043]
129. Struckmann N, Schwering S, Wiegand S, Gschnell A, Yamada M, Kummer W, Wess J, Haberberger RV. (2003) Role of muscarinic receptor subtypes in the constriction of peripheral airways: studies on receptor-deficient mice. Mol Pharmacol, 64 (6): 1444-51. [PMID:14645675]
130. Sundaram K, Murugaian J, Watson M, Sapru H. (1989) M2 muscarinic receptor agonists produce hypotension and bradycardia when injected into the nucleus tractus solitarii. Brain Res, 477 (1-2): 358-62. [PMID:2467726]
131. Sykes DA, Dowling MR, Leighton-Davies J, Kent TC, Fawcett L, Renard E, Trifilieff A, Charlton SJ. (2012) The Influence of receptor kinetics on the onset and duration of action and the therapeutic index of NVA237 and tiotropium. J Pharmacol Exp Ther, 343 (2): 520-8. [PMID:22854200]
132. Takeuchi T, Fujinami K, Goto H, Fujita A, Taketo MM, Manabe T, Matsui M, Hata F. (2005) Roles of M2 and M4 muscarinic receptors in regulating acetylcholine release from myenteric neurons of mouse ileum. J Neurophysiol, 93 (5): 2841-8. [PMID:15574798]
133. Tanis SP, Plewe MB, Johnson TW, Butler SL, Dalvie D, DeLisle D, Dress KR, Hu Q, Huang B, Kuehler JE et al.. (2010) Azaindole N-methyl hydroxamic acids as HIV-1 integrase inhibitors-II. The impact of physicochemical properties on ADME and PK. Bioorg Med Chem Lett, 20 (24): 7429-34. [PMID:21036042]
134. ten Berge RE, Zaagsma J, Roffel AF. (1996) Muscarinic inhibitory autoreceptors in different generations of human airways. Am J Respir Crit Care Med, 154 (1): 43-9. [PMID:8680697]
135. Trendelenburg AU, Gomeza J, Klebroff W, Zhou H, Wess J. (2003) Heterogeneity of presynaptic muscarinic receptors mediating inhibition of sympathetic transmitter release: a study with M2- and M4-receptor-deficient mice. Br J Pharmacol, 138 (3): 469-80. [PMID:12569072]
136. Tränkle C, Dittmann A, Schulz U, Weyand O, Buller S, Jöhren K, Heller E, Birdsall NJ, Holzgrabe U, Ellis J et al.. (2005) Atypical muscarinic allosteric modulation: cooperativity between modulators and their atypical binding topology in muscarinic M2 and M2/M5 chimeric receptors. Mol Pharmacol, 68 (6): 1597-610. [PMID:16157694]
137. Tränkle C, Weyand O, Voigtländer U, Mynett A, Lazareno S, Birdsall NJ, Mohr K. (2003) Interactions of orthosteric and allosteric ligands with [3H]dimethyl-W84 at the common allosteric site of muscarinic M2 receptors. Mol Pharmacol, 64 (1): 180-90. [PMID:12815174]
138. Tseng J, Erbe CB, Kwitek AE, Jacob HJ, Popper P, Wackym PA. (2002) Radiation hybrid mapping of five muscarinic acetylcholine receptor subtype genes in Rattus norvegicus. Hear Res, 174 (1-2): 86-92. [PMID:12433399]
139. Tyagi S, Tyagi P, Van-le S, Yoshimura N, Chancellor MB, de Miguel F. (2006) Qualitative and quantitative expression profile of muscarinic receptors in human urothelium and detrusor. J Urol, 176 (4 Pt 1): 1673-8. [PMID:16952712]
140. Tzavara ET, Bymaster FP, Felder CC, Wade M, Gomeza J, Wess J, McKinzie DL, Nomikos GG. (2003) Dysregulated hippocampal acetylcholine neurotransmission and impaired cognition in M2, M4 and M2/M4 muscarinic receptor knockout mice. Mol Psychiatry, 8 (7): 673-9. [PMID:12874603]
141. Vakalopoulos A, Basting D, Brechmann M, Teller H, Boultadakis Arapinis M, Straub A, Mittendorf J, Meininghaus M, Müller T, Nowak-Reppel K et al.. (2024) Discovery of BAY 2413555, First Selective Positive Allosteric Modulator of the M2 Receptor to Restore Cardiac Autonomic Balance. J Med Chem, 67 (21): 19165-19187. [PMID:39463278]
142. Valant C, Felder CC, Sexton PM, Christopoulos A. (2012) Probe dependence in the allosteric modulation of a G protein-coupled receptor: implications for detection and validation of allosteric ligand effects. Mol Pharmacol, 81 (1): 41-52. [PMID:21989256]
143. Valuskova P, Farar V, Forczek S, Krizova I, Myslivecek J. (2018) Autoradiography of 3H-pirenzepine and 3H-AFDX-384 in Mouse Brain Regions: Possible Insights into M1, M2, and M4 Muscarinic Receptors Distribution. Front Pharmacol, 9: 124. [PMID:29515448]
144. Vilaró MT, Palacios JM, Mengod G. (1994) Multiplicity of muscarinic autoreceptor subtypes? Comparison of the distribution of cholinergic cells and cells containing mRNA for five subtypes of muscarinic receptors in the rat brain. Brain Res Mol Brain Res, 21 (1-2): 30-46. [PMID:8164520]
145. Wackym PA, Chen CT, Ishiyama A, Pettis RM, López IA, Hoffman L. (1996) Muscarinic acetylcholine receptor subtype mRNAs in the human and rat vestibular periphery. Cell Biol Int, 20 (3): 187-92. [PMID:8673067]
146. Waelbroeck M, De Neef P, Domenach V, Vandermeers-Piret MC, Vandermeers A. (1996) Binding of the labelled muscarinic toxin 125I-MT1 to rat brain muscarinic M1 receptors. Eur J Pharmacol, 305 (1-3): 187-92. [PMID:8813552]
147. Wang SZ, el-Fakahany EE. (1993) Application of transfected cell lines in studies of functional receptor subtype selectivity of muscarinic agonists. J Pharmacol Exp Ther, 266 (1): 237-43. [PMID:7687290]
148. Watson J, Brough S, Coldwell MC, Gager T, Ho M, Hunter AJ, Jerman J, Middlemiss DN, Riley GJ, Brown AM. (1998) Functional effects of the muscarinic receptor agonist, xanomeline, at 5-HT1 and 5-HT2 receptors. Br J Pharmacol, 125 (7): 1413-20. [PMID:9884068]
149. Weiner DM, Levey AI, Brann MR. (1990) Expression of muscarinic acetylcholine and dopamine receptor mRNAs in rat basal ganglia. Proc Natl Acad Sci USA, 87 (18): 7050-4. [PMID:2402490]
150. Wess J. (2003) Novel insights into muscarinic acetylcholine receptor function using gene targeting technology. Trends Pharmacol Sci, 24 (8): 414-20. [PMID:12915051]
151. Wess J. (2004) Muscarinic acetylcholine receptor knockout mice: novel phenotypes and clinical implications. Annu Rev Pharmacol Toxicol, 44: 423-50. [PMID:14744253]
152. Wess J, Duttaroy A, Zhang W, Gomeza J, Cui Y, Miyakawa T, Bymaster FP, McKinzie L, Felder CC, Lamping KG et al.. (2003) M1-M5 muscarinic receptor knockout mice as novel tools to study the physiological roles of the muscarinic cholinergic system. Recept Channels, 9 (4): 279-90. [PMID:12893539]
153. Wess J, Lambrecht G, Mutschler E, Brann MR, Dörje F. (1991) Selectivity profile of the novel muscarinic antagonist UH-AH 37 determined by the use of cloned receptors and isolated tissue preparations. Br J Pharmacol, 102 (1): 246-50. [PMID:2043926]
154. Widzowski DV, Bialobok P, Kucera KE, Mihut R, Sitar S, Knowles M, Stagnitto M, Howell A, McCreedy S, Machulskis A, Gordon J, Marler M, Wu ESC, Mullen G, Triggle DJ, Blosser J. (1997) Development of a pharmacological target profile for muscarinic agonists. Drug Development Research, 40: 117-132.
155. Wood MD, Murkitt KL, Ho M, Watson JM, Brown F, Hunter AJ, Middlemiss DN. (1999) Functional comparison of muscarinic partial agonists at muscarinic receptor subtypes hM1, hM2, hM3, hM4 and hM5 using microphysiometry. Br J Pharmacol, 126 (7): 1620-4. [PMID:10323594]
156. Yamashita Y, Tanaka K, Asano T, Yamakawa N, Kobayashi D, Ishihara T, Hanaya K, Shoji M, Sugai T, Wada M et al.. (2014) Synthesis and biological comparison of enantiomers of mepenzolate bromide, a muscarinic receptor antagonist with bronchodilatory and anti-inflammatory activities. Bioorg Med Chem, 22 (13): 3488-97. [PMID:24844758]
157. Yasuda RP, Ciesla W, Flores LR, Wall SJ, Li M, Satkus SA, Weisstein JS, Spagnola BV, Wolfe BB. (1993) Development of antisera selective for m4 and m5 muscarinic cholinergic receptors: distribution of m4 and m5 receptors in rat brain. Mol Pharmacol, 43 (2): 149-57. [PMID:8429821]
158. Zhang Q, Pacheco MA, Doupnik CA. (2002) Gating properties of GIRK channels activated by Galpha(o)- and Galpha(i)-coupled muscarinic m2 receptors in Xenopus oocytes: the role of receptor precoupling in RGS modulation. J Physiol (Lond.), 545 (2): 355-73. [PMID:12456817]
159. Zhang W, Basile AS, Gomeza J, Volpicelli LA, Levey AI, Wess J. (2002) Characterization of central inhibitory muscarinic autoreceptors by the use of muscarinic acetylcholine receptor knock-out mice. J Neurosci, 22 (5): 1709-17. [PMID:11880500]
160. Zhang X, Hernandez MR, Yang H, Erickson K. (1995) Expression of muscarinic receptor subtype mRNA in the human ciliary muscle. Invest Ophthalmol Vis Sci, 36 (8): 1645-57. [PMID:7541396]
161. Zhou C, Fryer AD, Jacoby DB. (2001) Structure of the human M(2) muscarinic acetylcholine receptor gene and its promoter. Gene, 271 (1): 87-92. [PMID:11410369]