
Hot topics in pharmacology archive
2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007
2019
June 2019
Predicting and Understanding the Human Microbiome's Impact on Pharmacology
(1) Hitchings R & Kelly L. (2019). Predicting and Understanding the Human Microbiome's Impact on Pharmacology. Trends Pharmacol Sci,pii: S0165-6147(19)30091-4. doi: 10.1016/j.tips.2019.04.014. [PMID: 31171383]
The Complement Receptor C5aR2: A Powerful Modulator of Innate and Adaptive Immunity
(1) Li XX et al. (2019). The Complement Receptor C5aR2: A Powerful Modulator of Innate and Adaptive Immunity. J Immunol, 202(12):3339-3348. doi: 10.4049/jimmunol.1900371. [PMID: 31160390]
Understanding ligand binding selectivity in a prototypical GPCR family
(1) Mattedi G et al. (2019). Understanding ligand binding selectivity in a prototypical GPCR family. J Chem Inf Model, doi: 10.1021/acs.jcim.9b00298. [PMID: 31125224]
Functional characterization of 3D protein structures informed by human genetic diversity
(1) Hicks M et al. (2019). Functional characterization of 3D protein structures informed by human genetic diversity. PNAS USA, 116(18):8960-8965. doi: 10.1073/pnas.1820813116. [PMID: 30988206]
Cryo-EM structure of oxysterol-bound human Smoothened coupled to a heterotrimeric Gi
(1) Qi X et al. (2019). Cryo-EM structure of oxysterol-bound human Smoothened coupled to a heterotrimeric Gi. Nature, doi: 10.1038/s41586-019-1286-0. [PMID: 31168089]
PRECOG: PREdicting COupling probabilities of G-protein coupled receptors
(1) Singh G et al. (2019). PRECOG: PREdicting COupling probabilities of G-protein coupled receptors. Nucleic Acids Res, pii: gkz392. doi: 10.1093/nar/gkz392. [PMID: 31143927]
Capturing mixture composition: an open machine-readable format for representing mixed substances
(1) Clark AM et al. (2019). Capturing mixture composition: an open machine-readable format for representing mixed substances. J Cheminform, 11(1):33. doi: 10.1186/s13321-019-0357-4. [PMID: 31124006]
Drug-Target Association Kinetics in Drug Discovery
(1) IJzermann AP & Guo D (2019). Drug-Target Association Kinetics in Drug Discovery. Trends Biochem Sci, S0968-0004(19)30084-2. doi: 10.1016/j.tibs.2019.04.004. [PMID: 31101454]
May 2019
Depressive disorders: Treatment failures and poor prognosis over the last 50 years
(1) Warne T et al. (2019). Depressive disorders: Treatment failures and poor prognosis over the last 50 years. Pharmacol Res Perspect, 7(3):e00472. doi: 10.1002/prp2.472. [PMID: 31072904]
No Support for Historical Candidate Gene or Candidate Gene-by-Interaction Hypotheses for Major Depression Across Multiple Large Samples
(1) Border R et al. (2019). No Support for Historical Candidate Gene or Candidate Gene-by-Interaction Hypotheses for Major Depression Across Multiple Large Samples. Am J Psychiatry, 176(5):376-387. doi: 10.1176/appi.ajp.2018.18070881. [PMID: 30845820]
There is no "Depression Gene"
(1) Lowe D. (2019). In The Pipeline: There is no "Depression Gene". Sci Trans Med, 10 May 2019. [In The Pipeline: Article]
Time to FRET about GPCR activation dynamics?
Comments by Shane D Hellyer and Karen J Gregory, Drug Discovery Biology, Monash Institute of Pharmaceutical Sciences, Monash University, Australia
G protein coupled receptors (GPCRs) are crucial for the transduction of extracellular stimuli to the intracellular space. Upon activation, GPCRs undergo large conformational changes to engage transducers and stimulate intracellular responses. However, the kinetics of agonist induced GPCR conformational changes are relatively understudied. An exception to this is the class A rhodopsin receptor, which has a covalent agonist and fast (< 1ms) activation kinetics. In contrast, other GPCRs are thought to activate across the low to mid millisecond range [1]. For Class C GPCRs, which are distinct from class A receptors in that they contain large extracellular agonist binding domains and exist as obligate dimers, the site of agonist binding is >100Å from where the transducer interacts [2]. Class C GPCR activation involves both dimer rearrangement and activation of the 7-transmembrane (7-TM) domain, which are thought to occur over 20-200ms [3-5]. An outstanding question is whether the activation kinetics of rhodopsin are indeed faster than other GPCRs, or if previous experimental approaches lacked sufficient resolution to reveal fast kinetics in other receptor families. To this end, Grushevskyi and colleagues have used FRET recordings to detect submillisecond activation dynamics of a prototypical class C GPCR, metabotropic glutamate receptor subtype 1 (mGlu1), demonstrating that mGlu1 undergoes two temporally distinct conformational changes upon activation [6]. Read the full article on our blog 
(1) Lohse M.J. et al. (2012). Fluorescence/bioluminescence resonance energy transfer techniques to study G protein-coupled receptor activation and signalling. Pharmacol Rev, 64: 299-336. [PMIDs: 22407612]
(2) Leach K & Gregory K.J. (2017) Molecular insights into allosteric modulation of Class C G protein-coupled receptors. Pharmacol Res, 116: 105-118. [PMIDs: 27965032]
(3) Hlacvackova V et al. (2012) Sequential inter- and intrasubunit rearrangements during activation of dimeric metabotropic glutamate receptor 1. Sci Signal, 5: ra59. [PMIDs: 22894836]
(4) Marcaggi P et al. (2009) Optical measurement of mGluR1 conformational changes reveals fast activation, slow deactivation, and sensitization. PNAS, 106: 11388-11393. [PMIDs: 19549872]
(5) Vafabakhsh R et al. (2015) Conformational dynamics of a class C G-protein-coupled receptor. Nature, 524: 497-501. [PMIDs: 26258295]
(6) Grushevskyi E.O. et al. (2019) Stepwise activation of a class C GPCR begins with millisecond dimer rearrangement. PNAS. pii: 201900261. doi:10.1073/pnas.1900261116. [Epub ahead of print] [PMIDs: 31023886]
Molecular basis for high-affinity agonist binding in GPCRs
(1) Warne T et al. (2019). Molecular basis for high-affinity agonist binding in GPCRs. Science, pii: eaau5595. doi: 10.1126/science.aau5595. [PMID: 31072904]
Aminergic GPCR-Ligand Interactions: A Chemical and Structural Map of Receptor Mutation Data
(1) Vass M et al. (2019). Aminergic GPCR-Ligand Interactions: A Chemical and Structural Map of Receptor Mutation Data. J Med Chem, 62(8):3784-3839. doi: 10.1021/acs.jmedchem.8b00836. [PMID: 30351004]
Fixing the Unfixable: The Art of Optimizing Natural Products for Human Medicine
(1) Yñigez-Guitierrez AR & Bachmann BO (2019). Fixing the Unfixable: The Art of Optimizing Natural Products for Human Medicine. J Med Chem, doi: 10.1021/acs.jmedchem.9b00246. [PMID: 31026161]
April 2019
Structural basis of ligand recognition at the human MT1 melatonin receptor
(1) Stauch B et al. (2019). Structural basis of ligand recognition at the human MT1 melatonin receptor. Nature, https://doi.org/10.1038/s41586-019-1141-3. [Nature: Article]
XFEL structures of the human MT2 melatonin receptor reveal the basis of subtype selectivity
(1) Johansson LC et al. (2019). XFEL structures of the human MT2 melatonin receptor reveal the basis of subtype selectivity. Nature, https://doi.org/10.1038/s41586-019-1144-0. [Nature: Article]
Analysis of tractable allosteric sites in G protein-coupled receptors
(1) Wakefield AE et al. (2019). Analysis of tractable allosteric sites in G protein-coupled receptors. Sci Rep, 9(1):6180. doi: 10.1038/s41598-019-42618-8. [PMID:30992500]
Small Molecule Allosteric Modulators of G-Protein-Coupled Receptors: Drug-Target Interactions
(1) Lu S & Zhang J. (2019). Small Molecule Allosteric Modulators of G-Protein-Coupled Receptors: Drug-Target Interactions. J Med Chem, 62(1):24-45. doi: 10.1021/acs.jmedchem.7b01844. [PMID:29457894]
Uncovering new disease indications for G-protein coupled receptors and their endogenous ligands
(1) Freudenberg JM et al. (2018). Uncovering new disease indications for G-protein coupled receptors and their endogenous ligands. BMC Bioinformatics, 19(1):345. doi: 10.1186/s12859-018-2392-y. [PMID:30285606]
Functional characterization of 3D protein structures informed by human genetic diversity
(1) Hicks M et al. (2019). Functional characterization of 3D protein structures informed by human genetic diversity. PNAS USA, pii: 201820813. doi: 10.1073/pnas.1820813116. [Epub ahead of print]. [PMID:30988206]
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens
(1) Behan FM et al. (2019). Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature, doi: 10.1038/s41586-019-1103-9. [PMID:30971826]
Structure and dynamics of the active human parathyroid hormone receptor-1
(1) Zhao LH et al. (2019). Structure and dynamics of the active human parathyroid hormone receptor-1. Science, 364(6436):148-153. doi: 10.1126/science.aav7942. [PMID:30975883]
Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM
(1) Zhao Y et al. (2019). Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science, pii: eaaw8250. doi: 10.1126/science.aaw8250. [PMID:30975770]
Rapid Deorphanization of Human Olfactory Receptors in Yeast
(1) Yasi EA et al. (2019). Rapid Deorphanization of Human Olfactory Receptors in Yeast. Biochemistry, doi: 10.1021/acs.biochem.8b01208. [PMID:30977365]
March 2019
No Pain, and No Worries?
(1) Lowe D. (2019). In The Pipeline: No Pain, and No Worries?. Sci Trans Med, 29 March 2019. [In The Pipeline: Article]
Microdeletion in a FAAH pseudogene identified in a patient with high anandamide concentrations and pain insensitivity
(1) Habib AM et al. (2019). Microdeletion in a FAAH pseudogene identified in a patient with high anandamide concentrations and pain insensitivity. British Journal of Anaesthesia, https://doi.org/10.1016/j.bja.2019.02.019. [SciDirect: Abstract]
The past, present and future of anti-malarial medicines
(1) Tse EG et al. (2019). The past, present and future of anti-malarial medicines. Malar J, 22;18(1):93. doi: 10.1186/s12936-019-2724-z. [PMID:30902052]
Biology must develop herd immunity against bad-actor molecules
(1) Plemper RK & Cox RM. (2018). Biology must develop herd immunity against bad-actor molecules. PLoS Pathog, 14(6):e1007038. doi: 10.1371/journal.ppat.1007038. [PMID:29953540]
Rise up against statistical significance, probably.
Comments by Alistair Mathie (@AlistairMathie), The Medway School of Pharmacy
A recent commentary in Nature has the provocative title “Retire Statistical Significance” [1 with a list of more than 800 signatories] and has been widely interpreted as a call for the entire concept of statistical significance to be abandoned. Closer reading of the commentary suggests that the main message of the paper is a call to stop the use of p values or confidence intervals in a categorical or binary sense in order to be absolute as to whether a result supports or refutes a scientific hypothesis. This remains a radical proposal but perhaps does not signal the end for statistical tests in biomedical research just yet. Read the full article on our blog 
(1) Amrhein V, Greenland S & McShane B. (2019). Scientists rise up against statistical significance. Nature, 567(7748):305-307. doi: 10.1038/d41586-019-00857-9. [PMID:30894741]
(2) Curtis MJ et al. (2019). Experimental design and analysis and their reporting: new guidance for publication in BJP. Br J Pharmacol, 172(14):3461-71. doi: 10.1111/bph.12856. [PMID:26114403]
(3) Curtis MJ et al. (2019). Experimental design and analysis and their reporting II: updated and simplified guidance for authors and peer reviewers. Br J Pharmacol, 175(7):987-993. doi: 10.1111/bph.14153. [PMID:29520785]
(4) Colquhoun D. (2019). An investigation of the false discovery rate and the misinterpretation of p-values. R Soc Open Sci, 1(3):140216. doi: 10.1098/rsos.140216. eCollection 2014 Nov. [PMID:26064558]
(5) Casadevall A. (2019). Duke University’s huge misconduct fine is a reminder to reward rigour. Nature, 568(7). [World View: Article]
A Brief Note About Alzheimer’s
(1) Lowe D. (2019). In The Pipeline: A Brief Note About Alzheimer’s. Sci Trans Med, 21 March 2019. [In The Pipeline: Article]
Pharma R&D Annual Review 2018
(1) Lloyd I. (2019). Pharma R&D Annual Review 2018. Citeline. [Citeline:Whitepaper]
February 2019
Exciting Times for Ion Channel Pharmacology
Comments by Alistair Mathie (@AlistairMathie) and Emma L. Veale (@Ve11Emma), The Medway School of Pharmacy
Whilst life is always exciting as an ion channel pharmacologist, the last few months have been particularly so, with a large number of publications showing structures of ion channels with regulatory molecules bound to them. In just the last month, the journal, Science, has published several such papers. Three of these concern voltage-gated sodium channels (NaV1.2,, NaV1.7) and the binding of potent and selective toxins from animals [1-3]. Another reveals the structure of the primary human cooling and menthol sensor channel TRPM8 (id:500) bound to synthetic cooling and menthol-like compounds [4]. In the most recent paper [5], Schewe and colleagues extend their outstanding work on selectivity-filter gating of K2P potassium (K) channels (Schewe et al. (2016). Cell. PMID: 26919430), to identify a binding site for negatively charged activators of these channels (styled the “NCA binding site”) Read the full article on our blog 
(1) Clairfeuille T et al. (2019). Structural basis of α-scorpion toxin action on Nav channels. Science, pii: eaav8573. doi: 10.1126/science.aav8573. [PMID:30733386]
(2) Shen H et al. (2019). Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins. Science, pii: eaaw2493. doi: 10.1126/science.aaw2493. [Epub ahead of print]. [PMID:30765606]
(3) Pan X et al. (2019). Molecular basis for pore blockade of human NaNa+ channel Nav1.2 by the μ-conotoxin KIIIA. Science, pii: eaaw2999. doi: 10.1126/science.aaw2999. [Epub ahead of print]. [PMID:30765605]
(4) Yin Y et al. (2019). Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel. Science, pii: eaav9334. doi: 10.1126/science.aav9334. [Epub ahead of print]. [PMID:30733385]
(5) Schewe M et al. (2019). A pharmacological master key mechanism that unlocks the selectivity filter gate in K+ channels. Science, 363(6429):875-880. doi: 10.1126/science.aav0569.. [PMID:30792303]
Ligand biological activity predicted by cleaning positive and negative chemical correlations
Comments by Anthony Davenport, IUPHAR/BPS Guide to PHARMACOLOGY, University of Cambridge
New machine learning algorithm for drug discovery that is twice as efficient as the industry standard and identified potential ligands for the M1 receptor, a potential target for the treatment of Alzheimer’s disease [1]. Read the full article on our blog 
(1) Lee AA et al. (2019). Ligand biological activity predicted by cleaning positive and negative chemical correlations. PNAS, https://doi.org/10.1073/pnas.1810847116. [Epub ahead of print]. [PNAS: Article]
Ligand biological activity predicted by cleaning positive and negative chemical correlations
(1) Lee AA et al. (2019). Ligand biological activity predicted by cleaning positive and negative chemical correlations. PNAS, https://doi.org/10.1073/pnas.1810847116. [Epub ahead of print]. [PNAS: Article]
Recent updates in the discovery and development of novel antimalarial drug candidates
(1) Okombo J & Chibale K (2019). Recent updates in the discovery and development of novel antimalarial drug candidates. MedChemComm, 9(3):437-453. doi: 10.1039/c7md00637c. [PMID:30108934]
A class of highly selective inhibitors bind to an active state of PI3Kγ
(1) Gangadhara G et al. (2019). A class of highly selective inhibitors bind to an active state of PI3Kγ. Nat Chem Biol, doi: 10.1038/s41589-018-0215-0. [Epub ahead of print]. [PMID:30718815]
3,3'-Disubstituted 5,5'-Bi(1,2,4-triazine) derivatives with Potent in vitro and in vivo Antimalarial Activity
(1) Xue L et al. (2019). 3,3'-Disubstituted 5,5'-Bi(1,2,4-triazine) derivatives with Potent in vitro and in vivo Antimalarial Activity. J Med Chem, doi: 10.1021/acs.jmedchem.8b01799. [Epub ahead of print]. [PMID:30715882]
Antibodies and venom peptides: new modalities for ion channels
(1) Cox MA et al. (2019). Antibodies and venom peptides: new modalities for ion channels. Nat Rev Drug Discov, doi: 10.1038/s41573-019-0013-8. [Epub ahead of print]. [PMID:30728472]
Choline acetyltransferase-expressing T cells are required to control chronic viral infection
(1) Cox MA et al. (2019). Choline acetyltransferase-expressing T cells are required to control chronic viral infection. Science, 363(6427):639-644. doi: 10.1126/science.aau9072. [PMID:30733420]
Separating host and microbiome contributions to drug pharmacokinetics and toxicity
(1) Zimmermann M et al. (2019). Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science, 363(6427). pii: eaat9931. doi: 10.1126/science.aat9931. [PMID:30733391]
Diverse GPCRs exhibit conserved water networks for stabilization and activation
(1) Venkatakrishnan AJ et al. (2019). Diverse GPCRs exhibit conserved water networks for stabilization and activation. PNAS USA, pii: 201809251. doi: 10.1073/pnas.1809251116. [Epub ahead of print]. [PMID:30728297]
The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease
(1) Rothhammer V & Quintana FJ (2019). The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat Rev Immunol, https://doi.org/10.1038/s41577-019-0125-8. [NatRevImmunol: Abstract]
Ca2+ allostery in PTH-receptor signaling
(1) White AD et al. (2019). Ca2+ allostery in PTH-receptor signaling. PNAS USA, pii: 201814670. doi: 10.1073/pnas.1814670116. [Epub ahead of print]. [PMID:30718391]
Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity
(1) Vriens K et al. (2019). Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nat Struct Mol Biol, doi: 10.1038/s41586-019-0904-1. [Epub ahead of print]. [PMID:30728499]
Structures of the 5-HT2A receptor in complex with the antipsychotics risperidone and zotepine
(1) Kimura KT et al. (2019). Structures of the 5-HT2A receptor in complex with the antipsychotics risperidone and zotepine. Nat Struct Mol Biol, 26(2):121-128. doi: 10.1038/s41594-018-0180-z. [PMID:30723326]
January 2019
Global Portrait of Protein Targets of Metabolites of the Neurotoxic Compound BIA 10-2474
(1) Huang Z et al. (2019). Global Portrait of Protein Targets of Metabolites of the Neurotoxic Compound BIA 10-2474. ACS Chem Biol, doi: 10.1021/acschembio.8b01097. [Epub ahead of print]. [PMID:30702848]
Promises, promises, and precision medicine
(1) Joyner MJ & Paneth N (2019). Promises, promises, and precision medicine. J Clin Invest, pii: 126119. doi: 10.1172/JCI126119. [Epub ahead of print]. [PMID:30688663]
Pharmacological inhibition of GPR4 remediates intestinal inflammation in a mouse colitis model
(1) Sanderlin EJ et al. (2019). Pharmacological inhibition of GPR4 remediates intestinal inflammation in a mouse colitis model. bioRxiv, doi: https://doi.org/10.1101/533174. [bioRxiv:Abstract]
Navigating around Patented Routes by Preserving Specific Motifs along Computer-Planned Retrosynthetic Pathways
(1) Molga K et al. (2019). Navigating around Patented Routes by Preserving Specific Motifs along Computer-Planned Retrosynthetic Pathways. Chem, https://doi.org/10.1016/j.chempr.2018.12.004. [Chem in press:Article]
Hit Dexter 2.0: Machine-Learning Models for the Prediction of Frequent Hitters
(1) Stork C et al. (2019). Hit Dexter 2.0: Machine-Learning Models for the Prediction of Frequent Hitters. J Chem Inf Model, doi: 10.1021/acs.jcim.8b00677. [Epub ahead of print]. [PMID:30624935]
An online resource for GPCR structure determination and analysis
Comments by David E. Gloriam, University of Copenhagen (@David_Gloriam)
To accelerate the determination of GPCR structures and to help assess the quality of the available templates based on the modifications and methods, a recent article in Nature Methods presents “An Online Resource for GPCR Structure Determination and Analysis” [1]. Read the full article on our blog 
(1) Munk C et al. (2019). An online resource for GPCR structure determination and analysis. J Med Chem, doi:10.1038/s41592-018-0302-x. [PMID:30664776]
Fatty acid recognition in the Frizzled receptor family
(1) Nile AH et al. (2019). Fatty acid recognition in the Frizzled receptor family. J Biol Chem, 294(2):726-736. doi: 10.1074/jbc.REV118.005205. [PMID:30530496]
Hydroxamic acid inhibitors provide cross-species inhibition of Plasmodium M1 and M17 aminopeptidases
(1) Vinh NB et al. (2019). Hydroxamic acid inhibitors provide cross-species inhibition of Plasmodium M1 and M17 aminopeptidases. J Med Chem, doi: 10.1021/acs.jmedchem.8b01310. [Epub ahead of print]. [PMID:30537832]
2018 New Drug Therapy Approvals
(1) Woodcock J (2019). 2018 New Drug Therapy Approvals. FDA. [PDF:Report]
Secreted amyloid-β precursor protein functions as a GABA B R1a ligand to modulate synaptic transmission
(1) Rice HC et al. (2019). Secreted amyloid-β precursor protein functions as a GABA B R1a ligand to modulate synaptic transmission. Science, 363(6423). pii: eaao5213. doi: 10.1126/science.aao5213. [PMID:30630900]
What Makes a Kinase Promiscuous for Inhibitors?
(1) Hanson SM et al. (2018). What Makes a Kinase Promiscuous for Inhibitors? Cell Chem Biol, S2451-9456(18)30412-4. doi: 10.1016/j.chembiol.2018.11.005. [Epub ahead of print]. [PMID:30612951]
Using the drug-protein interactome to identify anti-ageing compounds for humans
(1) Fuentealba M et al. (2019). Using the drug-protein interactome to identify anti-ageing compounds for humans. PLoS comput Biol, 15(1):e1006639. doi: 10.1371/journal.pcbi.1006639. [Epub ahead of print]. [PMID:30625143]
Chronic TLR7 and TLR9 signaling drives anemia via differentiation of specialized hemophagocytes
(1) Akilesh HM et al. (2019). Chronic TLR7 and TLR9 signaling drives anemia via differentiation of specialized hemophagocytes. Science, 363(6423). pii: eaao5213. doi: 10.1126/science.aao5213. [PMID:30630901]
New Cannabinoid Receptors Structures
Comments by Steve Alexander, IUPHAR/BPS Guide to PHARMACOLOGY, (@mqzspa)
Cannabinoid receptors respond to multiple endogenous fatty acid derivatives and are often divided into neuronal-associated CB1 receptors and immune cell-associated CB2 receptors. Both receptors are GPCR, coupled predominantly to Gi, and have cytoprotective properties. The predominant psychotropic agent in Cannabis, THC, acts as a partial agonist at both receptors. CB1 patho/physiological responses are often characterised as analgesic, rewarding, orexigenic, hypothermic and amnestic, while CB2 receptors are mostly associated with anti-inflammatory effects. Two Cell papers [1,2] describe new structures for these receptors. Read the full article on our blog 
(1) Kumar K et al. (2018). Structure of a Signaling Cannabinoid Receptor 1-G Protein Complex. Cell, pii: S0092-8674(18)31565-4. doi: 10.1016/j.cell.2018.11.040. [Epub ahead of print]. [PMID:30639101]
(2) Liu X et al. (2018). Crystal Structure of the Human Cannabinoid Receptor CB2. Cell, pii: S0092-8674(18)31625-8. doi: 10.1016/j.cell.2018.12.011. [Epub ahead of print]. [PMID:30639103]
The IUPHAR Pharmacology Education Project
(1) Faccenda E et al. (2018). The IUPHAR Pharmacology Education Project. Clin Pharmacol Ther, 105(1):45-48. doi: 10.1002/cpt.1278. [PMID:30588614]
A patent review on PD-1/PD-L1 antagonists: small molecules, peptides, and macrocycles (2015-2018)
(1) Shaabani S et al. (2018). A patent review on PD-1/PD-L1 antagonists: small molecules, peptides, and macrocycles (2015-2018). Expert Opin Ther Pat, 28(9):665-678. doi: 10.1080/13543776.2018.1512706. [PMID:30107136]
BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification
(1) Djoumbou-Feunang Y et al. (2019). BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification. J Chemoinform, 11(1):2. doi: 10.1186/s13321-018-0324-5. [PMID:30612223]
GABAA receptor signalling mechanisms revealed by structural pharmacology
(1) Masiulis S et al. (2019). GABAA receptor signalling mechanisms revealed by structural pharmacology. Nat Commun., doi: 10.1038/s41586-018-0832-5. [Epub ahead of print]. [PMID:30602790]
Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists
(1) Schöppe J et al. (2019). Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists. Nat Commun., 10(1):17. doi: 10.1038/s41467-018-07939-8. [PMID:30604743]
The signposts and winding roads to immunity and inflammation
(1) Kanneganti TD (2019). The signposts and winding roads to immunity and inflammation. Nat Rev Immunol., doi: 10.1038/s41577-018-0108-1. [Epub ahead of print]. [PMID:30602731]
The Next Generation of Immunotherapy for Cancer: Small Molecules Could Make Big Waves
(1) Kerr WG & Chisholm JD (2019). The Next Generation of Immunotherapy for Cancer: Small Molecules Could Make Big Waves. J Immunol., 202(1):11-19. doi: 10.4049/jimmunol.1800991. [PMID:30587569]
The X-ray crystal structure of human endothelin 1, a polypeptide hormone regulator of blood pressure
(1) McPherson A & Larson SB (2019). The X-ray crystal structure of human endothelin 1, a polypeptide hormone regulator of blood pressure. Acta. Cryst., 75(Pt 1):47-53. doi: 10.1107/S2053230X18016011. [PMID:30605125]
2018
December 2018
Evolution of PI3Kγ and δ Inhibitors for Inflammatory and Autoimmune Diseases
(1) Perry MWD et al. (2018). Evolution of PI3Kγ and δ Inhibitors for Inflammatory and Autoimmune Diseases. J Med Chem, doi: 10.1021/acs.jmedchem.8b01298. [Epub ahead of print]. [PMID:30582813]
GPR37/GPR37L1 and the putative pairing with prosaptide/PSAP
Dr. Nicola J. Smith, Victor Chang Cardiac Research Institute, Australia
As is often the case with orphan GPCRs, assigning the endogenous ligand has been controversial for the closely related peptide family orphans, GPR37 and GPR37L1. In 2013, Randy Hall and his team (PubMed: 23690594) first reported an association between both centrally-expressed orphan GPCRs and prosaposin (PSAP) and prosaptide (TX14A), the synthetic active epitope of PSAP. Since that time there has been much debate in the field about whether this pairing is correct, with some authors corroborating the findings (PubMed: 24371137; 30010619, 28795439) and others not (PubMed: 23396314; 27072655; 28688853). Note that Head Activator, found in Hydra, was earlier reported as a ligand (PubMed: 16443751) but was quickly discredited (PubMed:28688853; 23686350).
A recent paper by Sergey Kasparov’s laboratory in Bristol has added further fuel to the fire. In a series of well controlled experiments, Liu et al. [1] provided convincing evidence that prosaptide is cyto- and neuro-protective and promotes chemotaxis. They are also the first group to demonstrate an effect of prosaptide at a more physiologically plausible potency. At the same time, Bang et al. [2] published a ground-breaking paper linking GPR37 expression to macrophage function. Moreover, they proposed a second, more potent ligand for GPR37 (GPR37L1 was not studied): the pro-resolving mediator neuroprotectin D1 (NPD1). Using HEK293 cells expressing GPR37, NPD1 was a potent stimulator of Gαi/o-dependent calcium flux; findings that were corroborated in macrophages isolated from wild type, but not GPR37 knock-out, mice (PubMed: 30010619). Thus, it may be that the endogenous ligand for GPR37 (and perhaps GPR37L1?) is not a peptide after all, but a lipid ... Read the full article on our blog 
(1) Liu B et al. (2018). Glio- and neuro-protection by prosaposin is mediated by orphan G-protein coupled receptors GPR37L1 and GPR37. Glia, 66(11):2414-2426. doi: 10.1002/glia.23480. [PMID:30260505]
(2) Bang S et al. (2018). GPR37 regulates macrophage phagocytosis and resolution of inflammatory pain. J Clin Invest, 128(8):3568-3582. doi: 10.1172/JCI99888. [PMID:30010619]
Improving the Gene Ontology Resource to Facilitate More Informative Analysis and Interpretation of Alzheimer's Disease Data
(1) Kramarz B et al. (2018). Improving the Gene Ontology Resource to Facilitate More Informative Analysis and Interpretation of Alzheimer's Disease Data. Genes (Basel), 9(12). pii: E593. doi: 10.3390/genes9120593. [PMID:30501127]
Structures shed light on prostanoid signaling
(1) Hollenstein K. (2018). Structures shed light on prostanoid signaling. Nat Chem Biol, 15(1):3-5. doi: 10.1038/s41589-018-0178-1. [PMID:30510191]
Systematic polypharmacology and drug repurposing via an integrated L1000-based Connectivity Map database mining
(1) Liu T-P et al. (2018). Systematic polypharmacology and drug repurposing via an integrated L1000-based Connectivity Map database mining. Royal Society Open Science, doi: https://doi.org/10.1098/rsos.181321. [Royal Society Open Science:Article]
QSAR-Based Virtual Screening: Advances and Applications in Drug Discovery
(1) Neves BJ et al. (2018).QSAR-Based Virtual Screening: Advances and Applications in Drug Discovery. Frontiers in Pharmacology, doi:https://doi.org/10.3389/fphar.2018.01275. [Frontiers:Mini Review Article]
Trend Analysis of a Database of Intravenous Pharmacokinetic Parameters in Humans for 1352 Drug Compounds
(1) Lombardo F et al. (2018). Trend Analysis of a Database of Intravenous Pharmacokinetic Parameters in Humans for 1352 Drug Compounds. Drug Metab Dispos, 46(11):1466-1477. doi: 10.1124/dmd.118.082966. [PMID:30115648]
Web-Based Tools for Polypharmacology Prediction
(1) Awale M & Reymond JL (2018). Web-Based Tools for Polypharmacology Prediction. Methods Mol Biol, 1888:255-272. doi: 10.1007/978-1-4939-8891-4_15. [PMID:30519952]
Open-source discovery of chemical leads for next-generation chemoprotective antimalarials
(1) Antonova-Koch Y et al. (2018). Open-source discovery of chemical leads for next-generation chemoprotective antimalarials. Science, 362(6419). doi:10.1126/science.aat9446. [Science:Article]
The convergence of artificial intelligence and chemistry for improved drug discovery
(1) Wright SC et al. (2018). The convergence of artificial intelligence and chemistry for improved drug discovery. Future Med Chem, doi: 10.4155/fmc-2018-0161. [Epub ahead of print]. [PMID:30499699]
FZD5 is a Gαq-coupled receptor that exhibits the functional hallmarks of prototypical GPCRs
(1) Wright SC et al. (2018). FZD5 is a Gαq-coupled receptor that exhibits the functional hallmarks of prototypical GPCRs. Sci Signal, 11(559). pii: eaar5536. doi: 10.1126/scisignal.aar5536. [PMID:30514810]
Machine learning in chemoinformatics and drug discovery
(1) Lo CY et al. (2018). Machine learning in chemoinformatics and drug discovery. Drug Discov Today, 23(8):1538-1546. doi: 10.1016/j.drudis.2018.05.010. [PMID:29750902]
Structural basis for ligand recognition of the human thromboxane A2 receptor
(1) Fan H et al. (2018). Structural basis for ligand recognition of the human thromboxane A2 receptor. Nat Chem Biol, 15(1):27-33. doi: 10.1038/s41589-018-0170-9. [PMID:30510189]
Chemical space of naturally occurring compounds
(1) Saldivar-Gonzalez FI et al. (2018). Chemical space of naturally occurring compounds. Physical Sciences Reviews, doi: https://doi.org/10.1515/psr-2018-0103. [Link:Abstract]
Organic synthesis in a modular robotic system driven by a chemical programming language
(1) Steiner S et al. (2018). Organic synthesis in a modular robotic system driven by a chemical programming language. Science, pii: eaav2211. doi: 10.1126/science.aav2211. [Epub ahead of print]. [PMID:30498165]
The drug repurposing landscape from 2012 to 2017: evolution, challenges, and possible solutions
(1) Polamreddy P & Gattu N (2018). The drug repurposing landscape from 2012 to 2017: evolution, challenges, and possible solutions. Drug Discov Today, pii: S1359-6446(18)30282-4. doi: 10.1016/j.drudis.2018.11.022. [Epub ahead of print]. [PMID:30513339]
November 2018
Genome-wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function
(1) Shifrut E et al. (2018). Genome-wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function. Cell, pii: S0092-8674(18)31333-3. doi: 10.1016/j.cell.2018.10.024. [PMID:30449619]
Doing it All - How Families are Reshaping Rare Disease Research
(1) Ekins S & Peristein EO (2018). Doing it All - How Families are Reshaping Rare Disease Research. Pharm Res, 35(10):192. doi: 10.1007/s11095-018-2481-7. [PMID:30116974]
Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states
(1) Manolaridis et al. (2018). Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states. Nature, 563(7731):426-430. doi: 10.1038/s41586-018-0680-3. [PMID:30405239]
Conformational ensemble of the human TRPV3 ion channel
(1) Zubcevic L et al. (2018). Conformational ensemble of the human TRPV3 ion channel. Nat Commun, 9(1):4773. doi: 10.1038/s41467-018-07117-w. [PMID:30429472]
Binding Kinetics Survey of the Drugged Kinome
(1) Georgi V et al. (2018). Binding Kinetics Survey of the Drugged Kinome. J Am Chem Soc, 140(46):15774-15782. doi: 10.1021/jacs.8b08048. [PMID:30362749]
Somatic APP gene recombination in normal and Alzheimer’s disease neurons
Comments by Jerold Chun, Sanford Burnham Prebys Medical Discovery Institute
A new facet of the human brain has been reported [1] involving a first example of somatic gene recombination in neurons, representing a normal neural mechanism whose disruption could underlie the most common (sporadic) forms of Alzheimer’s disease. Mosaic and somatic recombination of Amyloid Precursor Protein (APP) was identified in this well-known Alzheimer’s disease gene, where increased copies and mutations in rare families or Down syndrome are considered causal... Read the full article on our blog 
(1) Lee MH et al. (2018). Somatic APP gene recombination in Alzheimer’s disease and normal neurons. Nature, doi: 10.1038/s41586-018-0718-6. [Epub ahead of print]. [PMID:30464338]
Cellular thermal shift assays to measure ligand-to-target engagement
Comments by Dr. Thomas Lundbäck, Associate Director, Mechanistic Biology & Profiling, Discovery Sciences, AstraZeneca R&D, Gothenburg, Sweden
The cellular thermal shift assay (CETSA) was introduced in July of 2013 as a means to investigate drug target engagement inside live cells and tissues. The simplicity of CETSA has allowed prompt adoption in the literature but the importance of rapid changes in ligand binding is still not well recognised. To explore these considerations we systematically varied both the heat-pulse temperature and duration in CETSA using p38a as our model system [1]... Read the full article on our blog 
(1) Seashore-Ludlow B et al. (2018). Quantitative Interpretation of Intracellular Drug Binding and Kinetics Using the Cellular Thermal Shift Assay. Biochemistry, doi: 10.1021/acs.biochem.8b01057. [Epub ahead of print]. [PMID:30418016]
Assembly of a pan-genome from deep sequencing of 910 humans of African descent
(1) Sherman RM et al. (2018). Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat Genet, doi: 10.1038/s41588-018-0273-y. [Epub ahead of print]. [PMID:30455414]
Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation
(1) Visnes T (2018). Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation. Science, 362(6416):834-839. doi: 10.1126/science.aar8048. [PMID:30442810]
Be open about drug failures to speed up research
(1) Alteri E & Guizzaro L (2018). Be open about drug failures to speed up research. Nature, 563(7731):317-319. doi: 10.1038/d41586-018-07352-7. [PMID:30425369]
Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017-Utility and Limitations (Jan 2018)
(1) Baell JB & Nissick JWM (2018). Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017-Utility and Limitations. ACS Chem Biol, 13(1):36-44. doi: 10.1021/acschembio.7b00903. [PMID:29202222]
Genome Analyses of >200,000 Individuals Identify 58 Loci for Chronic Inflammation and Highlight Pathways that Link Inflammation and Complex Disorders
(1) Ligthart S et al. (2018). Genome Analyses of >200,000 Individuals Identify 58 Loci for Chronic Inflammation and Highlight Pathways that Link Inflammation and Complex Disorders. Am J Hum genet, 103(5):691-706. doi: 10.1016/j.ajhg.2018.09.009. [PMID:30388399]
Nanopore native RNA sequencing of a human poly(A) transcriptome
(1) Workman RE et al. (2018). Nanopore native RNA sequencing of a human poly(A) transcriptome. bioRxiv, doi: https://doi.org/10.1101/460410. [bioRxiv:Abstract]
Increased Obesity Is Causal for Increased Inflammation—A Mendelian Randomisation Study
(1) van Zuydam N et al. (2018). Increased Obesity Is Causal for Increased Inflammation—A Mendelian Randomisation Study. Diabetes, late breaking poster session, 67(supp 1), doi: https://doi.org/10.2337/db18-217-LB. [Diabetes:Abstract]
Crystal structure of human endothelin ETB receptor in complex with peptide inverse agonist IRL2500
(1) Nagiri C et al. (2018). Crystal structure of human endothelin ETB receptor in complex with peptide inverse agonist IRL2500. bioRxiv, doi: https://doi.org/10.1101/460410. [bioRxiv:Abstract]
The metabolite BH4 controls T cell proliferation in autoimmunity and cancer
(1) Cronin SJF et al. (2018). The metabolite BH4 controls T cell proliferation in autoimmunity and cancer. Nature, 563(7732):564-568. doi: 10.1038/s41586-018-0701-2. [PMID:30405245]
The signalling conformation of the insulin receptor ectodomain
(1) Weis F et al. (2018). The signalling conformation of the insulin receptor ectodomain. Nat Commun, 144:244-255. 9(1):4420. doi: 10.1038/s41467-018-06826-6. [PMID:30356040]
Drug-receptor kinetics and sigma-1 receptor affinity differentiate clinically evaluated histamine H3 receptor antagonists
(1) Riddy DM et al. (2018). Drug-receptor kinetics and sigma-1 receptor affinity differentiate clinically evaluated histamine H3 receptor antagonists. Neuropharmacology, 144:244-255. doi: 10.1016/j.neuropharm.2018.10.028. [Epub ahead of print]. [PMID:30359639]
Joining Forces: The Chemical Biology-Medicinal Chemistry Continuum (Sept 2017)
(1) Plowright AT et al. (2018). Joining Forces: The Chemical Biology-Medicinal Chemistry Continuum. Chem Cell Biol, 24(9):1058-1065. doi: 10.1016/j.chembiol.2017.05.019. [PMID:28602761]
MoonDB 2.0: an updated database of extreme multifunctional and moonlighting proteins
(1) Ribeiro DM et al. (2018). MoonDB 2.0: an updated database of extreme multifunctional and moonlighting proteins. Nucleic Acids Res, doi: 10.1093/nar/gky1039. [Epub ahead of print]. [PMID:30371819]
October 2018
The Future of Computational Chemogenomics
(1) Jacoby E & Brown JB (2018). The Future of Computational Chemogenomics. Computational Chemogenomics, pp 425-450. [SpringerLink:Protocol]
Slc7a5 regulates Kv1.2 channels and modifies functional outcomes of epilepsy-linked channel mutations
(1) Baronas VA et al. (2018). Slc7a5 regulates Kv1.2 channels and modifies functional outcomes of epilepsy-linked channel mutations. Nat Commun, 9(1):4417. doi: 10.1038/s41467-018-06859-x. [PMID:30356053]
Piezo channels and mechano-transduction in sensory neurons
Comments by Alistair Mathie (@AlistairMathie) and Emma L. Veale (@Ve11Emma), The Medway School of Pharmacy
Piezo channels (Piezo1 and Piezo2) are excitatory ion channels which respond directly to a variety of forms of mechanical stimuli. Two recent papers describe some of the critical roles of Piezo channels in sensory neuron transduction (1, 2). In the first, Murthy et al. (1) demonstrate that Piezo2 mediates both inflammatory and nerve-injury sensitized mechanical pain in mice. In the second, Zeng et al. (2), show that both Piezo1 and Piezo2 are responsible for the baro-receptor reflex that regulates blood pressure and cardiac function. Read the full article on our blog 
(1) Murthy SE et al. (2018). The mechanosensitive ion channel Piezo2 mediates sensitivity to mechanical pain in mice. Sci Transl Med, 10(462). pii: eaat9897. doi: 10.1126/scitranslmed.aat9897. [PMID: 30305457].
(2) Zeng WZ et al. (2018). PIEZOs mediate neuronal sensing of blood pressure and the baroreceptor reflex. Science, 362(6413):464-467. doi: 10.1126/science.aau6324. [PMID: 30361375].
A comprehensive analysis of the usability and archival stability of omics computational tools and resources
(1) Mangul S et al. (2018). A comprehensive analysis of the usability and archival stability of omics computational tools and resources. bioRxiv, doi: https://doi.org/10.1101/452532. [bioRxiv:Abstract]
Gene expression variability across cells and species shapes innate immunity
(1) Hagai T et al. (2018). Gene expression variability across cells and species shapes innate immunity. Nature, doi: 10.1038/s41586-018-0657-2. [Epub ahead of print]. [PMID:30356220]
From the Human Cell Atlas to dynamic immune maps in human disease
(1) Adlung L & Amit I. (2018). From the Human Cell Atlas to dynamic immune maps in human disease. Nat Rev Immunol, 18(10):597-598. doi: 10.1038/s41577-018-0050-2. [PMID:30078033]
Two Decades under the Influence of the Rule of Five and the Changing Properties of Approved Oral Drugs
(1) Schultz MD. (2018). Two Decades under the Influence of the Rule of Five and the Changing Properties of Approved Oral Drugs. J Med Chem, doi: 10.1021/acs.jmedchem.8b00686. [Epub ahead of print]. [PMID:30212196]
Role of RTP type D on reward association with cocaine administration
Comments by Steve Alexander, IUPHAR/BPS Guide to PHARMACOLOGY, (@mqzspa)
This report [1] from the National Institute on Drug Abuse in the USA focusses on the role of RTP type D on reward associated with cocaine administration. They identify that RTP type D heterozygous knockout mice exhibit lower reward responses to cocaine, and that a novel small molecule that appears to target the enzymatic function of RTP type D is able to reduce cocaine-induced place preference and self-administration in wild type, but not heterozygous knockout mice. Read the full article on our blog 
(1) Uhl GR et al. (2018). Cocaine reward is reduced by decreased expression of receptor-type protein tyrosine phosphatase D (PTPRD) and by a novel PTPRD antagonist. Proc Natl Acad Sci USA, pii: 201720446. doi: 10.1073/pnas.1720446115. [Epub ahead of print]. [PMID: 30348770]
Genenames.org: the HGNC and VGNC resources in 2019
(1) Braschi B et al. (2018). Genenames.org: the HGNC and VGNC resources in 2019. Nucleic Acids Res, 5:180230. doi: 10.1093/nar/gky930. [Epub ahead of print]. [PMID:30304474]
A large-scale dataset of in vivo pharmacology assay results
(1) Swaminathan J et al. (2018). A large-scale dataset of in vivo pharmacology assay results. Nat Biotechnol, 5:180230. doi: 10.1038/nbt.4278. [Epub ahead of print]. [PMID:30346938]
A large-scale dataset of in vivo pharmacology assay results
(1) Hunter FMI et al. (2018). A large-scale dataset of in vivo pharmacology assay results. Sci Data, 5:180230. doi: 10.1038/sdata.2018.230. [PMID:30351302]
Structure of the human voltage-gated sodium channel Nav1.4 in complex with β1
(1) Pan X et al. (2018). Structure of the human voltage-gated sodium channel Nav1.4 in complex with β1. Science, 362(6412). pii: eaau2486. doi: 10.1126/science.aau2486. [PMID:30190309]
Rapid structure determination of microcrystalline molecular compounds using electron diffraction
(1) Gruene T et al. (2018). Rapid structure determination of microcrystalline molecular compounds using electron diffraction. Angew Chem Int Ed Eng, doi: 10.1002/anie.201811318. [Epub ahead of print]. [PMID:30325568]
Impact of Human Genetics on Drug R&D
(1) Plenge R (2018). Impact of Human Genetics on Drug R&D - Slideset. ASHG, doi: 10.1002/anie.201811318. [Epub ahead of print]. [link to PDF]
DRUG-seq for miniaturized high-throughput transcriptome profiling in drug discovery
(1) Ye C et al. (2018). DRUG-seq for miniaturized high-throughput transcriptome profiling in drug discovery. Nat Commun, 9(1):4307. doi: 10.1038/s41467-018-06500-x. [PMID:30333485]
Fibrin-targeting immunotherapy protects against neuroinflammation and neurodegeneration
(1) Ryu JK et al. (2018). Fibrin-targeting immunotherapy protects against neuroinflammation and neurodegeneration. Nat Immunol, 19(11):1212-1223. doi: 10.1038/s41590-018-0232-x. [PMID:30323343]
Drug repurposing: progress, challenges and recommendations
(1) Pushpakom S et al. (2018). Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov, doi: 10.1038/nrd.2018.168. [Epub ahead of print]. [PMID:30310233]
Regulatory T cells in the treatment of disease
(1) Sharabi A et al. (2018). Regulatory T cells in the treatment of disease. Nat Rev Drug Discov, 562(7726):181-183. doi: 10.1038/d41586-018-06956-3. [PMID:30310234]
The approach to predictive medicine that is taking genomics research by storm
(1) Warren M (2018). The approach to predictive medicine that is taking genomics research by storm. Nature, 562(7726):181-183. doi: 10.1038/d41586-018-06956-3. [PMID:30305759]
The UK Biobank resource with deep phenotyping and genomic data
(1) Bycroft C et al. (2018). The UK Biobank resource with deep phenotyping and genomic data. Nature, 62(7726):203-209. doi: 10.1038/s41586-018-0579-z. [PMID:30305743]
How Selective Are Pharmacological Inhibitors of Cell-Cycle-Regulating Cyclin-Dependent Kinases?
(1) Jorda R et al. (2018). How Selective Are Pharmacological Inhibitors of Cell-Cycle-Regulating Cyclin-Dependent Kinases? J Med Chem, 61(20):9105-9120. doi: 10.1021/acs.jmedchem.8b00049. [PMID:30234987]
Advanced model systems and tools for basic and translational human immunology
(1) Wager LE, DiFazio RM & Davis MM (2018). Advanced model systems and tools for basic and translational human immunology. Genome Med, 10(1):73. doi: 10.1186/s13073-018-0584-8. [PMID:30266097]
Where Do Recent Small Molecule Clinical Development Candidates Come From?
(1) Brown DG & Boström J (2018). Where Do Recent Small Molecule Clinical Development Candidates Come From? J Med Chem, doi: 10.1021/acs.jmedchem.8b00675. [Epub ahead of print]. [PMID:29920198]
Orally Active Peptides: Is There a Magic Bullet?
(1) Räder AFB et al. (2018). Orally Active Peptides: Is There a Magic Bullet? Agnew Chem Int Ed Eng, 57(44):14414-14438. doi: 10.1002/anie.201807298. [PMID:30144240]
LipidPedia: a comprehensive lipid knowledgebase
(1) Kuo TC & Tseng YJ (2018). LipidPedia: a comprehensive lipid knowledgebase. Bioinformatics, doi: 10.1093/bioinformatics/bty213. [Epub ahead of print]. [PMID:29648583]
Launching the C-HPP pilot project for functional characterization of identified proteins with no known function
(1) Paik YK et al. (2018). Launching the C-HPP pilot project for functional characterization of identified proteins with no known function. J Proteome Res, doi: 10.1021/acs.jproteome.8b00383. [Epub ahead of print]. [PMID:30269496]
International Union of Basic and Clinical Pharmacology. CVI: GABAA Receptor Subtype- and Function-selective Ligands: Key Issues in Translation to Humans
(1) Sieghart W & Savić MM (2018). International Union of Basic and Clinical Pharmacology. CVI: GABAA Receptor Subtype- and Function-selective Ligands: Key Issues in Translation to Humans. Pharmacol Rev, 70(4):836-878. doi: 10.1124/pr.117.014449. [PMID:30275042]
Uncovering new disease indications for G-protein coupled receptors and their endogenous ligands
(1) Freudenberg JM et al. (2018). Uncovering new disease indications for G-protein coupled receptors and their endogenous ligands. BMC Bioinformatics, 19(1):345. doi: 10.1186/s12859-018-2392-y. [PMID:30285606]
Structural basis for σ1 receptor ligand recognition
(1) Schmidt HR et al. (2018). Structural basis for σ1 receptor ligand recognition. Nat Struct Mol Biol, 25(10):981-987. doi: 10.1038/s41594-018-0137-2. [PMID:30291362]
The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis
(1) Janes J et al. (2018). The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis. Proc Natl Acad Sci USA, 115(42):10750-10755. doi: 10.1073/pnas.1810137115. [PMID:30282735]
September 2018
Accurate classification of BRCA1 variants with saturation genome editing
(1) Findlay GM et al. (2018). Accurate classification of BRCA1 variants with saturation genome editing. Nature, 562(7726):217-222. doi: 10.1038/s41586-018-0461-z. [PMID:30209399]
Structures of the Human PGD2 Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition
(1) Wang L et al. (2018). Structures of the Human PGD2 Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition. Mol Cell, pii: S1097-2765(18)30643-9. doi: 10.1016/j.molcel.2018.08.009. [Epub ahead of print]. [PMID:30220562]
Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits
(1) Evangelou E et al. (2018). Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet, doi: 10.1038/s41588-018-0205-x. [Epub ahead of print]. [PMID:30224653]
Discovery of a Potent, Long-Acting, and CNS-Active Inhibitor (BIA 10-2474) of Fatty Acid Amide Hydrolase
(1) Kiss LE et al. (2018). Discovery of a Potent, Long-Acting, and CNS-Active Inhibitor (BIA 10-2474) of Fatty Acid Amide Hydrolase. Chem Med Chem, doi: 10.1002/cmdc.201800393. [Epub ahead of print]. [PMID:30113139]
Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor
Comments by Steve Alexander, IUPHAR/BPS Guide to PHARMACOLOGY, (@mqzspa)
In this multi-author, multi-centre publication [1] lead by Denise Wootten and Patrick Sexton from the Monash Institute of Pharmacological Sciences, there is reported a 3.3 Angstrom structure of one of the more unusual G protein-coupled receptors. The CGRP receptor is a target for the recently FDA-approved monoclonal antibody erenumab targetting migraine. The receptor is unusual because of its modulation by a trio of accessory proteins exemplified here by RAMP1, which influence both the pharmacological and signalling profiles of the GPCR. Read the full article on our blog 
(1) Liang YL et al. (2018). Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor. Nature, doi: 10.1038/s41586-018-0535-y. [Epub ahead of print]. [PMID: 30175587]
Large-Scale Reanalysis of Publicly Available HeLa Cell Proteomics Data in the Context of the Human Proteome Project
(1) Robin T et al. (2018). Large-Scale Reanalysis of Publicly Available HeLa Cell Proteomics Data in the Context of the Human Proteome Project. J Proteome Res, doi: 10.1021/acs.jproteome.8b00392. [Epub ahead of print]. [PMID:30175587]
Therapeutic Targeting of Endosomal G-Protein-Coupled Receptors
(1) Thomsen ARB et al. (2018). Therapeutic Targeting of Endosomal G-Protein-Coupled Receptors. Trends Pharmacol Sci, pii: S0165-6147(18)30138-X. doi: 10.1016/j.tips.2018.08.003. [Epub ahead of print]. [PMID:30180973]
Expanding the medicinal chemistry synthetic toolbox
(1) Boström et al. (2018). Expanding the medicinal chemistry synthetic toolbox. Nat Rev Drug Discov, doi: 10.1038/nrd.2018.116. [Epub ahead of print]. [PMID:30140018]
Allosteric Modulation of Class A GPCRs: Targets, Agents, and Emerging Concepts
(1) Wold et al. (2018). Allosteric Modulation of Class A GPCRs: Targets, Agents, and Emerging Concepts. J Med Chem, doi: 10.1021/acs.jmedchem.8b00875. [Epub ahead of print]. [PMID:30106578]
August 2018
Artificial intelligence in drug discovery
(1) Sellwood et al. (2018). Artificial intelligence in drug discovery. J Chem Inf Model, 10(17):2025-2028. doi: 10.4155/fmc-2018-0212. [PMID:30101607]
Characterization of the Chemical Space of Known and Readily Obtainable Natural Products
(1) Chen Y et al. (2018). Characterization of the Chemical Space of Known and Readily Obtainable Natural Products. J Chem Inf Model, 58(8):1518-1532. doi: 10.1021/acs.jcim.8b00302. [PMID:30010333]
Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight
(1) Shi Y & Holtzman DM (2018). Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat Rev Immunol, doi: 10.1038/s41577-018-0051-1. [Epub ahead of print]. [PMID:30140051]
β-Subunit of the voltage-gated Ca2+ channel Cav1.2 drives signaling to the nucleus via H-Ras
Comments by Jörg Striessnig, University of Innsbruck
This paper [1] extends previous studies demonstrating a key role of voltage-gated L-type Ca2+ channels in the modulation of activity-dependent gene transcription. Earlier work in cultured neurons had already shown that L-type channel activity is required to activate gene expression through different signaling pathways, including the Ras/MAPK pathway (2-4). This is confirmed in the present study (primarily in experiments with HEK-293 cells) but there are important differences in the way nuclear signaling gets activated. Read the full article on our blog 
(1) Servili E et al. (2018). β-Subunit of the voltage-gated Ca2+ channel Cav1.2 drives signaling to the nucleus via H-Ras. Proc Natl Acad Sci USA, 115(37):E8624-E8633. doi: 10.1073/pnas.1805380115. [PMID: 30150369]
(2) Dolmetsch RE et al. (2001). Signaling to the nucleus by an L-type calcium channel-calmodulin complex through the MAP kinase pathway. Science, 294(5541):333-9. [PMID: 11598293]
(3) Wu GY et al. (2018). Activity-dependent CREB phosphorylation: convergence of a fast, sensitive calmodulin kinase pathway and a slow, less sensitive mitogen-activated protein kinase pathway. Cold Spring Harbor Perspect Biol, 98(5):2808-13. [PMID: 11226322]
(4) Hagenston AM & Bading H (2018). Calcium signaling in synapse-to-nucleus communication. Mol Pharmacol, 3(11):a004564. doi: 10.1101/cshperspect.a004564. [Epub ahead of print]. [PMID: 30021858]
Crystal structure of the Frizzled 4 receptor in a ligand-free state
(1) Yang S et al. (2018). Crystal structure of the Frizzled 4 receptor in a ligand-free state. Nature, 560(7720):666-670. doi: 10.1038/s41586-018-0447-x. [PMID:30135577]
Structural determinants of 5-HT2B receptor activation and biased agonism
(1) McCorvy JD et al. (2018). Structural determinants of 5-HT2B receptor activation and biased agonism.
Nat Struct Mol Biol, doi: 10.1038/s41594-018-0116-7. [Epub ahead of print]. [PMID:30127358]
Cryo-EM structures of the human volume-regulated anion channel LRRC8
(1) Kasuya G et al. (2018). Cryo-EM structures of the human volume-regulated anion channel LRRC8.
Nat Struct Mol Biol, doi: 10.1038/s41594-018-0109-6. [Epub ahead of print]. [PMID:30127360]
Mechanisms of signalling and biased agonism in G protein-coupled receptors
(1) Wootten D et al. (2018). Mechanisms of signalling and biased agonism in G protein-coupled receptors.
Nat Rev Mol Cell Biol, doi: 10.1038/s41580-018-0049-3. [Epub ahead of print]. [PMID:30104700]
Challenges of Connecting Chemistry to Pharmacology: Perspectives from Curating the IUPHAR/BPS Guide to PHARMACOLOGY
(1) Southan C et al. (2018). Challenges of Connecting Chemistry to Pharmacology: Perspectives from Curating the IUPHAR/BPS Guide to PHARMACOLOGY.
ACS Omega, 3(7):8408-8420. doi: 10.1021/acsomega.8b00884. [PMID:30087946]
PubChem chemical structure standardization
(1) Hähnke VD et al. (2018). PubChem chemical structure standardization.
J Cheminform., 10(1):36. doi: 10.1186/s13321-018-0293-8. [PMID:30097821]
Polypharmacology by Design: A Medicinal Chemist's Perspective on Multitargeting Compounds
(1) Proschak E et al. (2018). Polypharmacology by Design: A Medicinal Chemist's Perspective on Multitargeting Compounds.
J Med Chem, doi: 10.1021/acs.jmedchem.8b00760. [Epub ahead of print]. [PMID:30035545]
Co-regulatory networks of human serum proteins link genetics to disease
(1) Emilsson V et al. (2018). Co-regulatory networks of human serum proteins link genetics to disease.
Science, pii: eaaq1327. doi: 10.1126/science.aaq1327. [Epub ahead of print]. [PMID:30072576]
July 2018
Direct activation of neuronal KV7 channels by GABA and gabapentin – a novel mechanism for reducing neuronal excitability
Comments by Emma L. Veale (@Ve11Emma) and Alistair Mathie (@AlistairMathie)
The potassium channels KV7.2-KV7.5 (KCNQ2-5; GtoPdb target IDs 561-564) regulate neuronal excitability in the mammalian nervous system. The best characterised neuronal KV7 channels give rise to the M current and are mediated predominantly by hetero-tetramers of KV7.2 and KV7.3 subunits. Established anticonvulsant agents such as retigabine are known to dampen neuronal excitability by activating neuronal KV7 channels. A tryptophan in the S5 transmembrane region of neuronal KV7 channels is essential for retigabine sensitivity with KV7.3 channels particularly sensitive. Importantly, this residue is not present in the cardiac KV7.1 (KCNQ1) channel, reducing the potential for cardiac side effects.
Now, a pair of studies (1, 2) has shown that this same region of the channel also contributes to a high affinity binding site for GABA and related metabolites. Read the full article on our blog 
(1) Manville RW et al. (2018). Direct neurotransmitter activation of voltage-gated potassium channels. Nat Commun, 9(1):1847. doi: 10.1038/s41467-018-04266-w. [PMID: 29748663]
(2) Manville RW et al. (2018). Gabapentin is a potent activator of KCNQ3 and KCNQ5 potassium channels. Mol Pharmacol, pii: mol.118.112953. doi: 10.1124/mol.118.112953. [Epub ahead of print]. [PMID: 30021858]
All Eyes on Biopharma Trends
(1) Sutton S. (2018). All Eyes on Biopharma Trends.
The Medicine Maker, July 2018. [Full text:Medicine Maker]
TreeGrafter: phylogenetic tree-based annotation of proteins with Gene Ontology terms and other annotations
(1) Tang H, Finn RD & Thomas PD (2018). TreeGrafter: phylogenetic tree-based annotation of proteins with Gene Ontology terms and other annotations.
Bioinformatics, doi: 10.1093/bioinformatics/bty625. [Epub ahead of print]. [PMID:30032202]
VCE-004.3, A Cannabidiol Aminoquinone Derivative, Prevents Bleomycin-induced Skin Fibrosis and Inflammation Through PPARγ AND CB2 -dependent Pathways
(1) Del Rio C et al. (2018). VCE-004.3, A Cannabidiol Aminoquinone Derivative, Prevents Bleomycin-induced Skin Fibrosis and Inflammation Through PPARγ AND CB2 -dependent Pathways.
Br J Pharmacology, doi: 10.1111/bph.14450. [Epub ahead of print]. [PMID:30033591]
A rare loss-of-function variant of ADAM17 is associated with late-onset familial Alzheimer's disease
(1) Hartl D et al. (2018). A rare loss-of-function variant of ADAM17 is associated with late-onset familial Alzheimer disease.
Mol Psychiatry, doi: 10.1038/s41380-018-0091-8. [Epub ahead of print]. [PMID:29988083]
The Assay Guidance Manual: Quantitative Biology and Pharmacology in Preclinical Drug Discovery
(1) Coussens NP et al. (2018). The Assay Guidance Manual: Quantitative Biology and Pharmacology in Preclinical Drug Discovery.
Clin Transl Sci, doi: 10.1111/cts.12570. [Epub ahead of print]. [PMID:29877628]
Loose ends: almost one in five human genes still have unresolved coding status
(1) Abascal F et al. (2018). Loose ends: almost one in five human genes still have unresolved coding status.
Nucleic Acids Res, doi: 10.1093/nar/gky587. [Epub ahead of print]. [PMID:29982784]
Whole genome sequencing identifies high-impact variants in well-known pharmacogenomic genes
(1) Choi J, Tantisira K & Duan Q (2018). Whole genome sequencing identifies high-impact variants in well-known pharmacogenomic genes.
bioRxiv, doi: https://doi.org/10.1101/368225. [bioRxiv:Abstract]
Modeling polypharmacy side effects with graph convolutional networks
(1) Zitnik M et al. (2018). Modeling polypharmacy side effects with graph convolutional networks.
Bioinformatics, 34(13):i457-i466. doi: 10.1093/bioinformatics/bty294. [PMID:29949996]
Cryo-EM structure of the human neutral amino acid transporter ASCT2
(1) Garaeva AA et al. (2018). Cryo-EM structure of the human neutral amino acid transporter ASCT2.
Nat Struct Mol Biol, 25(6):515-521. doi: 10.1038/s41594-018-0076-y. [PMID:29872227]
5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology
(1) Peng Y et al. (2018). 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172(4):719-730.e14. doi: 10.1016/j.cell.2018.01.001. [PMID:29398112]
Diverse antimalarials from whole-cell phenotypic screens disrupt malaria parasite ion and volume homeostasis
(1) Dennis ASM et al. (2018). Diverse antimalarials from whole-cell phenotypic screens disrupt malaria parasite ion and volume homeostasis.
Sci Rep, 8(1):8795. doi: 10.1038/s41598-018-26819-1. [PMID:29892073]
June 2018
New pharmacological tools reiterate lack of direct connection between Angiotensin 1–7 and the MAS1 GPCR
Comments by Sadashiva S. Karnik (karniks@ccf.org.us) and Kalyan Tirupula
A new type of deorphanization conundrum confronted in pairing the GPCR, MAS1 with the hormonal peptide angiotensin 1–7 (Ang1-7) was emphasised in recent IUPHAR reviews (1, 2). More evidence for disconnection between Ang1-7 and MAS1 is presented in the recent paper by Gaidarov et al. (3). Ang1-7 is produced by ACE2 or neutral endopeptidase by cleavage of a single amino acid, phenylalanine 8, from angiotensin II (AngII), the major renin angiotensin system hormone. MAS1 was described as the primary receptor for Ang1-7 in regulating diverse biological activities, including vasodilatory, cardio-protective, antithrombotic, antidiuretic and antifibrotic effects (4). These activities are lost in tissues of MAS1-deficient animals, producing striking phenotypes observed in the cardiovascular, renovascular, nervous and reproductive systems. Vast physiological responses to Ang1-7 studied in MAS1-deficient animals serve as the most compelling argument in favor of Ang1-7 pairing with MAS1. However, support for a direct interaction of Ang1-7 with MAS1 lack demonstration of classical G protein signaling and desensitization response to Ang1-7, as well as a lack consensus on confirmatory molecular pharmacological analyses (1, 2). Read the full article on our blog 
(1) Gaidarov I et al. (2018). Angiotensin (1–7) does not interact directly with MAS1, but can potently antagonize signaling from the AT1 receptor. Cell Signal, 50:9-24. doi: 10.1016/j.cellsig.2018.06.007. [Epub ahead of print]. [PMID: 29928987]
Biased Ligands of G Protein-Coupled Receptors (GPCRs): Structure-Functional Selectivity Relationships (SFSRs) and Therapeutic Potential
(1) Tan L et al. (2018). Biased Ligands of G Protein-Coupled Receptors (GPCRs): Structure-Functional Selectivity Relationships (SFSRs) and Therapeutic Potential.
J Med Chem, doi: 10.1021/acs.jmedchem.8b00435. [Epub ahead of print]. [PMID:29939744]
Structure of a human synaptic GABAA receptor
(1) Zhu S et al. (2018). Structure of a human synaptic GABAA receptor.
Nature, 559(7712):67-72. doi: 10.1038/s41586-018-0255-3. [PMID:29950725]
Structure of the adenosine-bound human adenosine A1 receptor–Gi complex
Comments by Steve Alexander, IUPHAR/BPS Guide to PHARMACOLOGY, (@mqzspa)
The A1 adenosine receptor is, for most people, a molecular target they can become conscious of when they block it, which happens frequently. Rapid consumption of higher doses of caffeine, in products like Italian espresso or Turkish coffee, provokes a rapid, transient increase in heart rate and a noticeable increase in limb tremor. In this report, a 3.6 Å structure of the receptor complexed with the endogenous agonist, adenosine, in the presence of the heterotrimeric G12 protein has been resolved by cryo-EM. Read the full article on our blog 
(1) Draper-Joyce CJ et al. (2018). Structure of the adenosine-bound human adenosine A1 receptor–Gi complex. Nature, 558, 559-563, doi: 10.1038/s41586-018-0236-6. [Epub ahead of print]. [PMID: 29925945]
Illuminating GPCR Signaling by Cryo-EM
(1) Safdari HA et al. (2018). Illuminating GPCR Signaling by Cryo-EM.
Trends in Cell Biology, doi.org/10.1016/j.tcb.2018.06.002. [Trends In Cell Biology:Summary]
Analysis of shared heritability in common disorders of the brain
(1) Brainstorm Consortium (2018). Analysis of shared heritability in common disorders of the brain.
Science, 360(6395). pii: eaap8757. doi: 10.1126/science.aap8757. [PMID:29930110]
Exploring Drugbank in Virtual Reality Chemical Space
(1) Probst D & Reymond J-L (2018). Exploring Drugbank in Virtual Reality Chemical Space. Science, doi.org/10.26434/chemrxiv.6629150.v1. [ChemRxiv:Abstract]
In vivo brain GPCR signaling elucidated by phosphoproteomics
(1) Liu JJ et al. (2018). In vivo brain GPCR signaling elucidated by phosphoproteomics. Science, 360(6395). pii: eaao4927. doi: 10.1126/science.aao4927. [PMID:29930108]
Recommendations toward a human pathway-based approach to disease research
(1) Marshall LJ et al. (2018). Recommendations toward a human pathway-based approach to disease research. Drug Discov Today, pii: S1359-6446(17)30473-7. doi: 10.1016/j.drudis.2018.05.038. [Epub ahead of print]. [PMID:29870792]
Defining Inflammatory Cell States in Rheumatoid Arthritis Joint Synovial Tissues by Integrating Single-cell Transcriptomics and Mass Cytometry
(1) Zhang F et al. (2018). Defining Inflammatory Cell States in Rheumatoid Arthritis Joint Synovial Tissues by Integrating Single-cell Transcriptomics and Mass Cytometry. BioRxiv, doi.org/10.1101/351130. [bioRxiv:Abstract]
Structural details for coupling of the agonist-occupied μ opioid receptor (amongst others) to the Gi protein
Comments by Eamonn Kelly & Katy Sutcliffe, University of Bristol
Every few years in the field of receptor pharmacology, a technological advance occurs that drives the field forward in terms of insight and understanding. Over the past couple of years, the cryo-EM technique (the development of which won the 2017 Nobel Prize in Chemistry for Dubochet, Frank, and Henderson) for resolving protein structures at near atomic resolution has been highlighted as one such approach. Now some of the first papers applying this methodology to G protein-coupled receptors (GPCRs) are beginning to appear Read the full article on our blog 
(1) Garcia-Nafria J et al. (2018). Cryo-EM structure of the serotonin 5-HT1B receptor coupled to heterotrimeric Go. Nature, 558(7711):620-623. doi: 10.1038/s41586-018-0241-9. [PMID:29925951]
Systems Medicine, Disease Maps and the future of Systems Biology
Comments by Steve Watterson, University of Ulster, (@systemsbiology)
What will Systems Biology look like in the future? Up to now, it has focussed on the development of standards, software tools and databases that enabled us to study the dynamics of physiological function mechanistically. However as these tools and technologies have matured, the focus of the systems biology research community has moved towards how they can best be interconnected and exploited to develop our understanding of health and disease across whole cells, tissues, organs, organisms. This version 2.0 of systems biology, will build on the existing technologies to create resources that are more intuitive, more accurate, more accessible and are easier to use for anyone engaged with research. Read the full article on our blog 
(1) Mazein M et al. (2018). Systems medicine disease maps: community-driven comprehensive representation of disease mechanisms. NPJ Syst Biol Appl., 4:21. doi: 10.1038/s41540-018-0059-y. eCollection 2018. [PMID:29872544]
Immune-centric network of cytokines and cells in disease context identified by computational mining of PubMed
(1) Kveler K et al. (2018). Immune-centric network of cytokines and cells in disease context identified by computational mining of PubMed. Nat Biotechnol., doi: 10.1038/nbt.4152. [Epub ahead of print]. [PMID:29912209]
Crystal structure of the µ-opioid receptor bound to a morphinan antagonist
(1) Manglik A et al. (2018). Crystal structure of the µ-opioid receptor bound to a morphinan antagonist. Nature, 485(7398):321-6. doi: 10.1038/nature10954. [PMID:22437502]
Emerging Approaches for the Identification of Protein Targets of Small Molecules - A Practitioners' Perspective
(1) Comess KM et al. (2018). Emerging Approaches for the Identification of Protein Targets of Small Molecules - A Practitioners' Perspective. J Med Chem, doi: 10.1021/acs.jmedchem.7b01921. [Epub ahead of print]. [PMID:29718665]
Targeted protein degradation and the enzymology of degraders
(1) Fischer SL & Phillips AJ (2018). Targeted protein degradation and the enzymology of degraders. Curr Opin Chem Biol, 44:47-55. doi: 10.1016/j.cbpa.2018.05.004. [Epub ahead of print]. [PMID:29885948]
The Assay Guidance Manual: Quantitative Biology and Pharmacology in Preclinical Drug Discovery
(1) Coussens NP et al. (2018). The Assay Guidance Manual: Quantitative Biology and Pharmacology in Preclinical Drug Discovery. Clin Transl Sci, doi: 10.1111/cts.12570. [Epub ahead of print]. [PMID:29877628]
Cryo-EM in drug discovery: achievements, limitations and prospects
(1) Renaud JP et al. (2018). Cryo-EM in drug discovery: achievements, limitations and prospects. Nat Rev Drug Discov, doi: 10.1038/nrd.2018.77. [Epub ahead of print]. [PMID:29880918]
Oxidized phospholipids are proinflammatory and proatherogenic in hypercholesterolaemic mice
(1) Que X et al. (2018). Oxidized phospholipids are proinflammatory and proatherogenic in hypercholesterolaemic mice. Nature, doi: 10.1038/s41586-018-0198-8. [Epub ahead of print]. [PMID:29875409]
Genomic atlas of the human plasma proteome
(1) Sun BB et al. (2018). Genomic atlas of the human plasma proteome. Nature, 558(7708):73-79. doi: 10.1038/s41586-018-0175-2. [PMID:29875488]
Clinical trial design and dissemination: comprehensive analysis of clinicaltrials.gov and PubMed data since 2005
(1) Zwierzyna M et al. (2018). Clinical trial design and dissemination: comprehensive analysis of clinicaltrials.gov and PubMed data since 2005. BMJ, doi: 10.1136/bmj.k2130. [BMJ:Article]
From genome-wide associations to candidate causal variants by statistical fine-mapping
(1) Schaid DJ, Chen W & Larson NB. (2018). From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat Rev Genet, doi: 10.1038/s41576-018-0016-z. [Epub ahead of print]. [PMID:29844615]
New Modalities, Technologies, and Partnerships in Probe and Lead Generation: enabling a Mode-of-Action centric paradigm
(1) Valuer E & Jimonet P. (2018). New Modalities, Technologies, and Partnerships in Probe and Lead Generation: enabling a Mode-of-Action centric paradigm. J Med Chem, doi: 10.1021/acs.jmedchem.8b00378. [Epub ahead of print]. [PMID:29851477]
How artificial intelligence is changing drug discovery
(1) Fleming N. (2018). How artificial intelligence is changing drug discovery. Nature, 557(7707):S55-S57. doi: 10.1038/d41586-018-05267-x. [PMID:29849160]
A big data approach to the concordance of the toxicity of pharmaceuticals in animals and humans
(1) Clark M & Steger-Hartmann T. (2018). A big data approach to the concordance of the toxicity of pharmaceuticals in animals and humans. Regul Toxicol Pharmacol, 96:94-105. doi: 10.1016/j.yrtph.2018.04.018. [PMID:29730448]
25 Years of Molecular Biology Databases: A Study of Proliferation, Impact, and Maintenance
(1) Imker HJ. (2018). 25 Years of Molecular Biology Databases: A Study of Proliferation, Impact, and Maintenance. Front. Res. Metr. Anal, doi: 10.3389/frma.2018.00018. [Full Article]
CypReact: A Software Tool for in Silico Reactant Prediction for Human Cytochrome P450 Enzymes
(1) Tian S et al. (2018). CypReact: A Software Tool for in Silico Reactant Prediction for Human Cytochrome P450 Enzymes. Future Med Chem, doi: 10.1021/acs.jcim.8b00035. [Epub ahead of print]. [PMID:29738669]
Advocating for mutually beneficial access to shelved compounds
(1) Pulley JM et al. (2018). Advocating for mutually beneficial access to shelved compounds. Future Med Chem, doi: 10.4155/fmc-2018-0090. [Epub ahead of print]. [PMID:29788759]
Structural basis for signal recognition and transduction by platelet-activating-factor receptor
(1) Cao C et al. (2018). Structural basis for signal recognition and transduction by platelet-activating-factor receptor. Nat Struc Mol Biol, 25(6):488-495. doi: 10.1038/s41594-018-0068-y. [PMID:29808000]
Integrating rare genetic variants into pharmacogenetic drug response predictions
(1) Ingelman-Sundberg M et al. (2018). Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum Genomics, 12(1):26. doi: 10.1186/s40246-018-0157-3. [PMID:29793534]
May 2018
Reciprocal signalling by Notch–Collagen V–CALCR retains muscle stem cells in their niche
(1) Baghdadi MB et al. (2018). Reciprocal signalling by Notch–Collagen V–CALCR retains muscle stem cells in their niche. Nature, doi: 10.1038/s41586-018-0144-9. [Nature:Abstract]
The Cellosaurus, a Cell-Line Knowledge Resource
(1) Bairoch A. (2018). The Cellosaurus, a Cell-Line Knowledge Resource. J Biomol Tech., doi: 10.7171/jbt.18-2902-002 [Epub ahead of print]. [PMC:PMC5945021]
Genomic, Proteomic and Phenotypic Heterogeneity in HeLa Cells across Laboratories: Implications for Reproducibility of Research Results
(1) Liu Y et al. (2018). Genomic, Proteomic and Phenotypic Heterogeneity in HeLa Cells across Laboratories: Implications for Reproducibility of Research Results. bioRxiv., doi: 10.1101/307421. [bioRxiv:Abstract]
Evidence that TLR4 Is Not a Receptor for Saturated Fatty Acids but Mediates Lipid-Induced Inflammation by Reprogramming Macrophage Metabolism
(1) Lancaster GI et al. (2018). Evidence that TLR4 Is Not a Receptor for Saturated Fatty Acids but Mediates Lipid-Induced Inflammation by Reprogramming Macrophage Metabolism. Cell Metab., 27(5):1096-1110.e5. doi: 10.1016/j.cmet.2018.03.014. [PMID:29681442]
A Census of Disease Ontologies
(1) Haendel M et al. (2018). A Census of Disease Ontologies. Annual Reviews, doi: 10.1146/annurev-biodatasci-080917-013459. [Abstract]
Drugs as habitable planets in the space of dark chemical matter
(1) Siramshetty VB & Preissner R. (2018). Drugs as habitable planets in the space of dark chemical matter. Drug Discov Today, 23(3):481-486. doi: 10.1016/j.drudis.2017.07.003. [PMID:28709991]
Metabolism as a Target for Modulation in Autoimmune Diseases
(1) Huang N & Perl A. (2018). Metabolism as a Target for Modulation in Autoimmune Diseases. Trends Immunol., S1471-4906(18)30083-8. doi: 10.1016/j.it.2018.04.006. [Epub ahead of print]. [PMID:29739666]
Activation mechanisms for a universal signalling protein
(1) Krumm B & Roth BL (2018). Activation mechanisms for a universal signalling protein. Nature News and Views. doi: 10.1038/d41586-018-04977-6. [Article]
Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein
Comments by Steve Alexander, IUPHAR/BPS Guide to PHARMACOLOGY, (@mqzspa)
The A2A adenosine receptor is densely expressed in dopamine-rich areas of the brain and in the vasculature. It is the target of an adjunct medication for Parkinson’s Disease, istradefylline in Japan, an A2A receptor antagonist. The A2A adenosine receptor is an example of a Gs-coupled receptor, activation of which in the cardiovascular system leads to inhibition of platelet aggregation and vasorelaxation. This new report highlights the link between the receptor and the G protein to focus on areas of unexpected flexibility in the ligand binding region. Read the full article on our blog 
(1) Garcia-Nafría J et al. (2018). Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. eLife, 7. pii: e35946. doi: 10.7554/eLife.35946. [PMID: 29726815]
Emerging Paradigm of Intracellular Targeting of G Protein-Coupled Receptors
(1) Chaturvedi M et al. (2018). Emerging Paradigm of Intracellular Targeting of G Protein-Coupled Receptors. Trends in Biomedical Sci., [Epub ahead of print]. doi: 10.1016/j.tibs.2018.04.003. [Abstract]
Pharma’s broken business model — Part 2: Scraping the barrel in drug discovery
(1) Stott K (2018). IDO inhibitors appear to have wiped out. Endpoint News - Biotech Voices, 2nd May 2018. [Blog]
Pharma’s broken business model Part 1: An industry on the brink of terminal decline
(1) Stott K (2017). Pharma’s broken business model Part 1: An industry on the brink of terminal decline. Endpoint News - Biotech Voices, 28th Nov 2017. [Blog]
Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis
(1) Zhang M et al. (2018). Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science, 360(6388): pii: eaap7847. doi: 10.1126/science.aap7847. [PMID:29724925]
A double-wammy for BACE1 inhibitors
Comments by Chris Southan, IUPHAR/BPS Guide to PHARMACOLOGY, (@cdsouthan)
BACE1 (beta secretase 1, BACE-1 or BACE) has been a key target for Alzheimer's disease (AD) for nearly two decades (1). However, there was a major disappointment when the Phase III trials with the Merck inhibitor verubecestat failed unequivocally despite lowering A-beta levels. The termination is reported both in NCT01739348 and the May 2018 full paper on the trial results (2).
Read the full article on our blog 
(1) Egan MF et al. (2018). Randomized Trial of Verubecestat for Mild-to-Moderate Alzheimer's Disease. N Engl J Med., 378(18):1691-1703. doi: 10.1056/NEJMoa1706441. [PMID:29719179]
Conformational plasticity in the selectivity filter of the TRPV2 ion channel
Comments by Steve Alexander, IUPHAR/BPS Guide to PHARMACOLOGY, (@mqzspa)
The TRPV2 ion channel is the less well-characterised relative of the TRPV1 or vanilloid receptor that is activated by capsaicin. TRPV2 channels have many similarities to the TRPV1 channels, in that they are homotetrameric and respond to some of the same ligands (natural products such as cannabinoids) as well as being triggered at elevated temperatures. This study (1) focusses on a different common feature of the whole Transient Receptor Potential family, which are often described as non-selective cation channels. Read the full article on our blog 
(1) Zubcevic L et al. (2018). Conformational plasticity in the selectivity filter of the TRPV2 ion channel. Nat Struc Mol Biol., 25:405-415. doi:10.1038/s41594-018-0059-z. [Abstract]
IDO inhibitors appear to have wiped out
(1) Lowe D (2018). IDO inhibitors appear to have wiped out. STM: In The Pipeline, 1st May 2018. [Blog]
A TRP channel trio mediates acute noxious heat sensing
(1) Vandewauw I et al. (2018). A TRP channel trio mediates acute noxious heat sensing. Nature, 555(7698):662-666. doi: 10.1038/nature26137. [PMID:29539642]
A computationally driven analysis of the polyphenol-protein interactome
(1) Lacroix S et al. (2018). A computationally driven analysis of the polyphenol-protein interactome. Sci Rep., 8(1):2232. doi: 10.1038/s41598-018-20625-5. [PMID:29396566]
April 2018
Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector
(1) Wang C et al. (2018). Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector. Nat Med., [Epub ahead or print]. doi: 10.1038/s41591-018-0004-z. [PMID:29632371]
New Paradigms in Adenosine Receptor Pharmacology: Allostery, Oligomerization and Biased Agonism
(1) Vecchio EA et al. (2018). New Paradigms in Adenosine Receptor Pharmacology: Allostery, Oligomerization and Biased Agonism. Br J Pharmacol., [Epub ahead or print]. doi: 10.1111/bph.14337. [PMID:29679502]
Cryo-EM structure of substrate-bound human telomerase holoenzyme
(1) Nguyen THD et al. (2018). Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature, [Epub ahead or print]. doi: 10.1038/s41586-018-0062-x. [PMID:29695869]
Discovery of Selective RNA-Binding Small Molecules by Affinity-Selection Mass Spectrometry
(1) Rizvi NF et al. (2018). Discovery of Selective RNA-Binding Small Molecules by Affinity-Selection Mass Spectrometry. ACS Chem Biol., 13(3):820-831. doi: 10.1021/acschembio.7b01013. [PMID:29412640]
Structural basis for GPR40 allosteric agonism and incretin stimulation
(1) Ho JD et al. (2018). Structural basis for GPR40 allosteric agonism and incretin stimulation. Nat Commun., 9(1):1645. doi: 10.1038/s41467-017-01240-w. [PMID:29695780]
Can we accelerate medicinal chemistry by augmenting the chemist with Big Data and artificial intelligence?
(1) Griffen EJ et al. (2018). Can we accelerate medicinal chemistry by augmenting the chemist with Big Data and artificial intelligence? Drug Discov Today, [Epub ahead of print] pii: S1359-6446(17)30578-0. doi: 10.1016/j.drudis.2018.03.011. [PMID:29577971]
Medical relevance of protein-truncating variants across 337,205 individuals in the UK Biobank study
(1) DeBoever C et al. (2018). Medical relevance of protein-truncating variants across 337,205 individuals in the UK Biobank study. Nat Commun.,9(1):1612. doi: 10.1038/s41467-018-03910-9. [PMID:29691392]
The 100 000 Genomes Project: bringing whole genome sequencing to the NHS
(1) Turnbull C et al.The 100 000 Genomes Project: bringing whole genome sequencing to the NHS. BMJ,361:k1687. doi: 10.1136/bmj.k1687. [PMID:29691228]
Analysis of predicted loss-of-function variants in UK Biobank identifies variants protective for disease
(1) Emdin CA et al. (2018). Analysis of predicted loss-of-function variants in UK Biobank identifies variants protective for disease. Nat Commun.,9(1):1613. doi: 10.1038/s41467-018-03911-8. [PMID:29691411]
Are there physicochemical differences between allosteric and competitive ligands?
(1) Smith RD, Lu J & Carlson HA (2018). Are there physicochemical differences between allosteric and competitive ligands?. PLoS, 13(11):e1005813. doi: 10.1371/journal.pcbi.1005813. [PMID:29125840]
Redundancy in two major compound databases
(1) Yonchev D et al. (2018). Redundancy in two major compound databases. Drug Discov Today, [Epub ahead of print] pii: S1359-6446(18)30027-8. doi: 10.1016/j.drudis.2018.03.005. [PMID:29559364]
Donated chemical probes for open science
(1) Müller S et al.(2018). Donated chemical probes for open science. eLife, 7. pii: e34311. doi: 10.7554/eLife.34311.. [PMID:29676732]
PKIDB: A Curated, Annotated and Updated Database of Protein Kinase Inhibitors in Clinical Trials
(1) Carles F. et al.. (2018). PKIDB: A Curated, Annotated and Updated Database of Protein Kinase Inhibitors in Clinical Trials. Molecules, 23(4). pii: E908. doi: 10.3390/molecules23040908. [PMID:29662024]
Translating translation
(1) Austin CP. (2018). Translating translation. Nat Rev Drug Discov., [Epub ahead of print]. doi: 10.1038/nrd.2018.27. [PMID:29674698]
3D structure of the P2X3 receptor bound to a negative allosteric modifier, identifies a binding site that is a target for development of novel therapeutic agents
Comments by Dr. Charles Kennedy, University of Strathclyde
Negative allosteric modulators (NAMs) are of great interest in drug development because they offer improved scope for the production of receptor antagonists with enhanced subtype-selectivity. Indeed, many NAMs are already on the market or undergoing clinical trials. NAMs act by binding to sites within receptors that are distinct from the primary, orthosteric ligand binding site and can inhibit the structural rearrangements of a receptor that are induced by orthosteric agonist binding.
P2X receptors are ligand-gated cation channels for which ATP is the endogenous orthosteric agonist. They are expressed throughout the body and the evidence indicates that they have numerous functions, including in sympathetic and parasympathetic neurotransmission, perception of sound, taste and pain, and immune regulation. Seven P2X subunits have been identified, which form trimers, to produce at least twelve different receptor subtypes. A major issue within the field has been a lack of selective antagonists for most P2X subtypes. This is unsurprising given the amino acid sequence similarity within the ATP binding site. Several selective NAMs have now been developed, but little is known about where in receptors they act and how exactly they inhibit receptor activation.
AF-219 is small molecule NAM at P2X3 receptors that was reported to be effective in a phase II clinical trial for treatment of refractory chronic cough. Wang et al., (1) combined X-ray crystallography, molecular modelling, and mutagenesis, to identify the site and mode of action of AF-219. P2X3 receptors are composed of three subunits, each of which adopts a conformation that could be likened to the shape of a leaping dolphin. Read the full article on our blog 
(1) Wang J et al. (2018). Druggable negative allosteric site of P2X3 receptors. Proc Natl Acad Sci USA, [Epub ahead of print] pii: 201800907. doi: 10.1073/pnas.1800907115. [PMID:29674445]
Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms
(1) Liu TL et al. (2018). Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science, 360(6386):762-775. pii: eaaq1392. doi: 10.1126/science.aaq1392. [PMID:29674564]
GPR68 Senses Flow and Is Essential for Vascular Physiology
(1) Xu J et al. (2018). GPR68 Senses Flow and Is Essential for Vascular Physiology. Cell, 173(3):762-775. doi: 10.1016/j.cell.2018.03.076. [PMID:29677517]
Challenges in validating candidate therapeutic targets in cancer
(1) Settleman J, Sawyers CL & Hunter T (2018). Challenges in validating candidate therapeutic targets in cancer. eLife, 7. pii: e32402. doi: 10.7554/eLife.32402. [PMID:29417929]
Of mice, men and immunity: a case for evolutionary systems biology
(1) Ernst PB & Carvunis AR (2018). Of mice, men and immunity: a case for evolutionary systems biology. Nat Immunol., 19(5):421-425. doi: 10.1038/s41590-018-0084-4. [PMID:29670240]
High-Throughput Gene Expression Profiles to Define Drug Similarity and Predict Compound Activity
(1) De Wolf H et al. (2018). High-Throughput Gene Expression Profiles to Define Drug Similarity and Predict Compound Activity. Assay Drug Dev Technol., 16(3):162-176. doi: 10.1089/adt.2018.845. [PMID:29658791]
Structural basis of ligand binding modes at the neuropeptide Y Y1 receptor
(1) Yang Z et al. (2018). Structural basis of ligand binding modes at the neuropeptide Y Y1 receptor. Nature, 556(7702):520-524. doi: 10.1038/s41586-018-0046-x. [PMID:29670288]
Untangling Galectin-Driven Regulatory Circuits in Autoimmune Inflammation
(1) Toscano MA et al. (2018). Untangling Galectin-Driven Regulatory Circuits in Autoimmune Inflammation. Trends Mol Med., 24(4):348-363. doi: 10.1016/j.molmed.2018.02.008. [PMID:29555188]
Biocuration: Distilling data into knowledge
(1) International Society for Biocuration. (2018). Biocuration: Distilling data into knowledge. PLoS Biol., 16(4):e2002846. doi: 10.1371/journal.pbio.2002846. [PMID:29659566]
Interleukins and their signaling pathways in the Reactome biological pathway database
(1) Jupe S et al. (2018). Interleukins and their signaling pathways in the Reactome biological pathway database. J Allergy Clin Immunol., 141(4):1411-1416. doi: 10.1016/j.jaci.2017.12.992. [PMID:29378288]
Cryptic glucocorticoid receptor-binding sites pervade genomic NF-κB response elements
(1) Hudson WH et al. (2018). Cryptic glucocorticoid receptor-binding sites pervade genomic NF-κB response elements. Nat Commun., 9(1):1337. doi: 10.1038/s41467-018-03780-1. [PMID:29626214]
Identification of rare sequence variation underlying heritable pulmonary arterial hypertension
(1) Gräf S et al. (2018). Identification of rare sequence variation underlying heritable pulmonary arterial hypertension. Nat Commun., 9(1):1416. doi: 10.1038/s41467-018-03672-4. [PMID:29650961]
Accessing Expert‐Curated Pharmacological Data in the IUPHAR/BPS Guide to PHARMACOLOGY
(1) Sharman JL et al. (2018). Accessing Expert‐Curated Pharmacological Data in the IUPHAR/BPS Guide to PHARMACOLOGY. Curr Prot in Bionfomatics., 61(1):1.34.1-1.34.46. doi: 10.1002/cpbi.46. [Abstract]
Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity
(1) Pao KC et al. (2018). Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature, 556(7701):381-385. doi: 10.1038/s41586-018-0026-1. [PMID:29643511]
Innate immune memory in the brain shapes neurological disease hallmarks
(1) Wendeln AC et al. (2018). Innate immune memory in the brain shapes neurological disease hallmarks. Nature, 556(7701):332-328. doi: 10.1038/s41586-018-0023-4. [PMID:29643512]
Meta-analysis of genetic association with diagnosed Alzheimer's disease identifies novel risk loci and implicates Abeta, Tau, immunity and lipid processing
(1) Kunkle BW et al. (2018). Meta-analysis of genetic association with diagnosed Alzheimer's disease identifies novel risk loci and implicates Abeta, Tau, immunity and lipid processing. bioRxiv. doi: 10.1101/294629. [Abstract]
Chemical probes and drug leads from advances in synthetic planning and methodology
(1) Gerry CJ & Schreiber SL. (2018). Chemical probes and drug leads from advances in synthetic planning and methodology. Nat Rev Drug Discov., 17(5):333-352. doi: 10.1038/nrd.2018.53. [PMID:29651105]
The Proposal to Lower P Value Thresholds to .005.
(1) Ioannidis JPA. (2018). The Proposal to Lower P Value Thresholds to .005. JAMA, 319(14):1429-1430. doi: 10.1001/jama.2018.1536. [PMID:29566133]
A Library of Phosphoproteomic and Chromatin Signatures for Characterizing Cellular Responses to Drug Perturbations
(1) Litichevskiy L et al. (2018). A Library of Phosphoproteomic and Chromatin Signatures for Characterizing Cellular Responses to Drug Perturbations. Cell Syst., 6(4):424-443. doi: 10.1016/j.cels.2018.03.012. [PMID:29655704]
High-throughput mouse phenomics for characterizing mammalian gene function
(1) Brown SDM et al. (2018). High-throughput mouse phenomics for characterizing mammalian gene function. Nat Rev Genet., [Epub ahead of print]. doi: 10.1038/s41576-018-0005-2. [PMID:29626206]
Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes
(1) Mahajan A et al. (2018). Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat Genet., 50(4):559-571. doi: 10.1038/s41588-018-0084-1. [PMID:29632382]
Analysis of Human Sequence Data Reveals Two Pulses of Archaic Denisovan Admixture
(1) Browning SR et al. (2018). Analysis of Human Sequence Data Reveals Two Pulses of Archaic Denisovan Admixture. Cell, 173(1):53-61. doi: 10.1016/j.cell.2018.02.031. [PMID:29551270]
Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis
(1) Scapin G et al. (2018). Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis. Nature, 556(7699):122-125. doi: 10.1038/nature26153. [PMID:29512653]
Quantitative Prediction of Rate Constants for Aqueous Racemization To Avoid Pointless Stereoselective Syntheses
(1) Ballard A et al. (2018). Quantitative Prediction of Rate Constants for Aqueous Racemization To Avoid Pointless Stereoselective Syntheses. Angew Chem Int Ed Engl., 57(4):982-985. doi: 10.1002/anie.201709163. [PMID:29072355]
Searching and Extracting Data from the EMBL-EBI Complex Portal
(1) Meldal BHM, Orchard S. (2018). Searching and Extracting Data from the EMBL-EBI Complex Portal. Methods Mol Biol., 1764:377-390. doi: 10.1007/978-1-4939-7759-8_24. [PMID:29605928]
Accurate functional classification of thousands of BRCA1 variants with saturation genome editing
(1) Findlay GM et al. (2018). Accurate functional classification of thousands of BRCA1 variants with saturation genome editing. BioRxiv, doi: 10.1101/294520. [Epub ahead of print] [Abstract]
Opportunities and obstacles for deep learning in biology and medicine
(1) Ching T et al. (2018). Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface., 15(141). pii: 20170387. doi: 10.1098/rsif.2017.0387. [PMID:29618526]
The Immune Landscape of Cancer
(1) Thorsson V et al. (2018). The Immune Landscape of Cancer. Immunity, doi: 10.1016/j.immuni.2018.03.023. [Epub ahead of print] [Full text]
Disease-Causing Mutations in the G Protein Gαs Subvert the Roles of GDP and GTP
(1) Hu Q, Shokat KM. (2018). Disease-Causing Mutations in the G Protein Gαs Subvert the Roles of GDP and GTP. Cell, doi: 10.1016/j.cell.2018.03.018. [Epub ahead of print] [Full text]
Identification of Misclassified ClinVar Variants via Disease Population Prevalence
(1) Shah N et al. (2018). Identification of Misclassified ClinVar Variants via Disease Population Prevalence. Am J Hum Genet., 102(4):609-619. doi: 10.1016/j.ajhg.2018.02.019. [PMID:29625023]
Chemical Diversity in the G Protein-Coupled Receptor Superfamily
(1) Vass M et al. (2018). Chemical Diversity in the G Protein-Coupled Receptor Superfamily. Trends Pharmacol Sci., pii: S0165-6147(18)30035-X. doi: 10.1016/j.tips.2018.02.004. [Epub ahead of print] [PMID:29576399]
Synthesis and Characterization of a Bidirectional Photoswitchable Antagonist Toolbox for Real-Time GPCR Photopharmacology
(1) Hauwert NJ et al. (2018). Synthesis and Characterization of a Bidirectional Photoswitchable Antagonist Toolbox for Real-Time GPCR Photopharmacology. J Am Chem Soc., 140(12):4232-4243. doi: 10.1021/jacs.7b11422. [PMID:29470065]
Cross-disorder analysis of schizophrenia and 19 immune diseases reveals genetic correlation
(1) Pouget JG, et al. (2018). Cross-disorder analysis of schizophrenia and 19 immune diseases reveals genetic correlation. BioRxiv, doi: 10.1101/068684. [Epub ahead of print] [Abstract]
Planning chemical syntheses with deep neural networks and symbolic AI
(1) Segler MHS et al. (2018). Planning chemical syntheses with deep neural networks and symbolic AI. Nature, 555(7698):604-610. doi: 10.1038/nature25978. [PMID:29595767]
Biased signalling: from simple switches to allosteric microprocessors
(1) Smith JS et al. (2018). Biased signalling: from simple switches to allosteric microprocessors. Nat Rev Drug Discov, 17(4):243-260. doi: 10.1038/nrd.2017.229. [PMID:29302067]
ImmPort, toward repurposing of open access immunological assay data for translational and clinical research
(1) Bhattacharya S et al. (2018). ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data, 5:180015. doi: 10.1038/sdata.2018.15. [PMID:29485622]
Organic synthesis provides opportunities to transform drug discovery
(1) Blakemore DC et al. (2018). Organic synthesis provides opportunities to transform drug discovery. Nat Chem, 10(4):383-394. doi: 10.1038/s41557-018-0021-z. [PMID:29568051]
March 2018
3D structures of the closed acid-sensing ion channel (ASIC) shed light on the activation mechanism of these neuronal ion channels
Comments by Stephan Kellenberger, Université de Lausanne, Switzerland
ASICs are potential drug targets of interest. Their activation mechanism has however remained elusive. ASICs are neuronal, proton-gated, sodium-permeable channels that are expressed in the central and peripheral nervous system of vertebrates. They form a subfamily of the Epithelial Na channel / degenerin channel family, and contribute to pain sensation, fear, learning, and neurodegeneration after ischemic stroke. Depending on the extracellular pH, they exist in either one of three functional states: closed (resting), open and desensitized. While ASICs are at physiological pH 7.4 in the closed state, they open briefly upon extracellular acidification, before entering the non-conducting desensitized state. Crystal structures of the chicken ASIC1 channel in the desensitized and the open state were published several years ago. This structural information allowed, together with observations from functional studies, an understanding of the transitions between the open and the desensitized state. In contrast, the absence of structural information on the closed conformation of ASICs precluded so far a molecular understanding of their activation mechanism.
The Gouaux laboratory has now published structures of the homotrimeric chicken ASIC1 obtained at high pH by X-ray crystallography (2.95 Å resolution) and by single particle cryo-electron microscopy (3.7 Å) (1). Read the full article on our blog 
(1) Yoder N et al. (2018). Gating mechanisms of acid-sensing ion channels. Nature, 555(7696):397-401. doi: 10.1038/nature25782. [PMID:29513651]
Extensive impact of non-antibiotic drugs on human gut bacteria
(1) Maier L, et al. (2018). Extensive impact of non-antibiotic drugs on human gut bacteria. Nature, doi:10.1038/nature25979. [Epub ahead of print] [Full text]
A Machine Learning Approach Predicts Tissue-Specific Drug Adverse Events
(1) Madhukar NS, et al. (2018). A Machine Learning Approach Predicts Tissue-Specific Drug Adverse Events. BioRxiv, doi: 10.1101/288332. [Epub ahead of print] [Abstract]
Ranking Enzyme Structures in the PDB by Bound Ligand Similarity to Biological Substrates
(1) Tyzack JD et al. (2018). Ranking Enzyme Structures in the PDB by Bound Ligand Similarity to Biological Substrates. Structure, pii: S0969-2126(18)30049-2. doi: 10.1016/j.str.2018.02.009. [Epub ahead of print] [PMID:29551288]
Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel
(1) She J et al. (2018). Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel. Nature, doi: 10.1038/nature26139. [Epub ahead of print] [PMID:29562233]
Conflicting evidence for the role of JNK as a target in breast cancer cell proliferation: Comparisons between pharmacological inhibition and selective shRNA knockdown approaches
(1) Wood RA et al. (2018). Conflicting evidence for the role of JNK as a target in breast cancer cell proliferation: Comparisons between pharmacological inhibition and selective shRNA knockdown approaches. Pharmacol Res Perspect., 6(1). doi: 10.1002/prp2.376. [PMID:29417765]
Closely related, yet unique: Distinct homo- and heterodimerization patterns of G protein coupled chemokine receptors and their fine-tuning by cholesterol
(1) Gahbauer S et al. (2018). Closely related, yet unique: Distinct homo- and heterodimerization patterns of G protein coupled chemokine receptors and their fine-tuning by cholesterol. PLoS Comput Biol., 14(3):e1006062. doi: 10.1371/journal.pcbi.1006062. [PMID:29529028]
Engineered mini G proteins provide a useful tool for studying the activation of GPCRs in living cells
Comments by Shane C. Wright and Gunnar Schulte, Karolinska Institute
In order to stabilize the GPCR-G protein complex, an agonist must be bound to the receptor and the alpha subunit of the heterotrimer must be in a nucleotide-free state. Ground-breaking work by expert crystallographers made use of so-called mini G (mG) proteins to stabilize the active conformation of the adenosine A2A receptor in the presence of agonist and guanine nucleotides, but in the absence of Gβγ [1]. These engineered G proteins behave in a way that mimics the nucleotide-free state despite being bound to GDP; thus, they can be seen as conformational sensors of the active receptor state. This work paved the way for another study recently published in the Journal of Biological Chemistry led by Nevin A. Lambert that looked to build on this minimalistic approach to see if representative mG proteins from the four subclasses (Gs, Gi/o, Gq/11 and G12/13) could 1) detect active GPCRs and 2) retain coupling specificity [2]. Read the full article on our blog 
(1) Carpenter B et al. (2016) Structure of the adenosine A(2A) receptor bound to an engineered G protein. Nature, 536 (7614): 104-107. [PMID:27462812]
(2) Wan Q et al. (2018) Mini G protein probes for active G protein-coupled receptors (GPCRs) in live cells. J Biol Chem. pii: jbc.RA118.001975. doi: 10.1074/jbc.RA118.001975. [Epub ahead of print] [PMID:29523687]
Functionally distinct and selectively phosphorylated GPCR subpopulations co-exist in a single cell
(1) Shen A et al. (2018). Functionally distinct and selectively phosphorylated GPCR subpopulations co-exist in a single cell. Nat Commun., 9(1):1050. doi: 10.1038/s41467-018-03459-7. [PMID:29535304]
Heterologous Expression, Biosynthetic Studies, and Ecological Function of the Selective Gq-Signaling Inhibitor FR900359
(1) Crüsemann M et al. (2018). Heterologous Expression, Biosynthetic Studies, and Ecological Function of the Selective Gq-Signaling Inhibitor FR900359. Angew Chem Int Ed Engl., 57(3):836-840. doi: 10.1002/anie.201707996. [PMID:29194875]
A comprehensive and quantitative comparison of text-mining in 15 million full-text articles versus their corresponding abstracts
(1) Westergaard D et al. (2018). A comprehensive and quantitative comparison of text-mining in 15 million full-text articles versus their corresponding abstracts. PLoS Comput Biol., 4(2):e1005962. doi: 10.1371/journal.pcbi.1005962. [PMID:29447159]
Precision medicine screening using whole-genome sequencing and advanced imaging to identify disease risk in adults
(1) Perkins BA et al. (2018). Precision medicine screening using whole-genome sequencing and advanced imaging to identify disease risk in adults. Proc Natl Acad Sci U S A., pii: 201706096. doi: 10.1073/pnas.1706096114. [Epub ahead of print] [PMID:29555771]
Crystal structure of the human 5-HT1B serotonin receptor bound to an inverse agonist
(1) Yin W et al. (2018). Crystal structure of the human 5-HT1B serotonin receptor bound to an inverse agonist. Cell Discovery, 4. doi:10.1038/s41421-018-0009-2. [Epub ahead of print] [Full text]
Augmented Reality in Scientific Publications-Taking the Visualization of 3D Structures to the Next Level
(1) Wolle P et al. (2018). Augmented Reality in Scientific Publications-Taking the Visualization of 3D Structures to the Next Level. ACS Chem Biol., 13(3):496-499. doi: 10.1021/acschembio.8b00153. [PMID:29544257]
Exploring G Protein-Coupled Receptors (GPCRs) Ligand Space via Cheminformatics Approaches: Impact on Rational Drug Design
(1) Basith S et al. (2018). Exploring G Protein-Coupled Receptors (GPCRs) Ligand Space via Cheminformatics Approaches: Impact on Rational Drug Design. Frontiers in Pharmacology, doi:10.3389/fphar.2018.00128. [Epub ahead of print] [Full text]
WhichP450: a multi-class categorical model to predict the major metabolising CYP450 isoform for a compound
(1) Hunt PA et al. (2018). WhichP450: a multi-class categorical model to predict the major metabolising CYP450 isoform for a compound. J Comput Aided Mol Des., doi: 10.1007/s10822-018-0107-0. [Epub ahead of print] [PMID:29464466]
Validation of ligands in macromolecular structures determined by X-ray crystallography
(1) Smart OS et al. (2018). Validation of ligands in macromolecular structures determined by X-ray crystallography. Acta Crystallogr D Struct Biol., 74(Pt 3):228-236. doi: 10.1107/S2059798318002541. [PMID:29533230]
Translational Bioinformatics year in review 2018
(1) Altman R. (2018). [Translational Bioinformatics year in review 2018]
Pharma R&D Annual Review 2018
(1) Lloyd I. (2018). [Pharma R&D Annual Review 2018]
Small Molecule Interactome Mapping by Photoaffinity Labeling Reveals Binding Site Hotspots for the NSAIDs
(1) Gao J et al. (2018). Small Molecule Interactome Mapping by Photoaffinity Labeling Reveals Binding Site Hotspots for the NSAIDs. J Am Chem Soc., doi: 10.1021/jacs.7b11639. [Epub ahead of print] [PMID:29543447]
Kinase inhibitors: the road ahead
(1) Ferguson FM, Gray NS. (2018). Kinase inhibitors: the road ahead. Nat Rev Drug Discov., doi: 10.1038/nrd.2018.21. [Epub ahead of print] [PMID:29545548]
STRENDA DB: enabling the validation and sharing of enzyme kinetics data
(1) Swainston N et al. (2018). STRENDA DB: enabling the validation and sharing of enzyme kinetics data. FEBS J., doi: 10.1111/febs.14427. [Epub ahead of print] [PMID:29498804]
Genetic risk for Alzheimer's disease is concentrated in specific macrophage and microglial transcriptional networks
(1) Tansey KE et al. (2018). Genetic risk for Alzheimer's disease is concentrated in specific macrophage and microglial transcriptional networks. Genome Med., 10(1):14. doi: 10.1186/s13073-018-0523-8. [PMID:29482603]
Small Molecule Allosteric Modulators of G-Protein-Coupled Receptors: Drug-Target Interactions
(1) Lu S, Zhang J. (2018). Small Molecule Allosteric Modulators of G-Protein-Coupled Receptors: Drug-Target Interactions. J Med Chem, doi: 10.1021/acs.jmedchem.7b01844. [Epub ahead of print] [PMID:29457894]
Unexplored therapeutic opportunities in the human genome
Comments by Chris Southan, IUPHAR/BPS Guide to PHARMACOLOGY, @cdsouthan
Contemporary drug discovery is dominated by two related themes. The first of these is target validation upon which the sustainability of pharmaceutical R&D (in both the commercial and academic sectors) crucially depends. The second is the size of the pool of human proteins that are/could become tractable to being progressed towards clinical efficacy as their final validation step (otherwise known as the druggable proteome). This usefully detailed review, by a large team of authors, touches on both themes but with a focus on how the community might increase the target pool by data-driven knowledge expansion for hitherto less well characterised proteins [1]. Read the full article on our blog 
(1) Oprea TI et al. (2018). Unexplored therapeutic opportunities in the human genome. Nat Rev Drug Discov. doi: 10.1038/nrd.2018.14 [Epub ahead of print] [PMID:29472638]
Prevalence of clinical trial status discrepancies: A cross-sectional study of 10,492 trials registered on both ClinicalTrials.gov and the European Union Clinical Trials Register
(1) Fleminger J, Goldacre B. (2018). Prevalence of clinical trial status discrepancies: A cross-sectional study of 10,492 trials registered on both ClinicalTrials.gov and the European Union Clinical Trials Register. PLoS One, 13(3):e0193088. doi: 10.1371/journal.pone.0193088. [PMID:29513684]
An Augmented Pocketome: Detection and Analysis of Small-Molecule Binding Pockets in Proteins of Known 3D Structure
(1) Bhagavat R et al. (2018). An Augmented Pocketome: Detection and Analysis of Small-Molecule Binding Pockets in Proteins of Known 3D Structure. Structure, 26(3):499-512.e2. doi: 10.1016/j.str.2018.02.001. [PMID:29514079]
Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms
(1) Deng Z et al. (2018). Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms. Nat Struct Mol Biol., 25(3):252-260. doi: 10.1038/s41594-018-0037-5. [PMID:29483651]
The role of gut microbiome and associated metabolome in the regulation of neuroinflammation in multiple sclerosis and its implications in attenuating chronic inflammation in other inflammatory and autoimmune disorders
(1) Dopkins N, Nagarkatti PS, Nagarkatti M. (2018). The role of gut microbiome and associated metabolome in the regulation of neuroinflammation in multiple sclerosis and its implications in attenuating chronic inflammation in other inflammatory and autoimmune disorders. Immunology, doi: 10.1111/imm.12903. [Epub ahead of print] [PMID:29392733]
The rise of deep learning in drug discovery
(1) Chen H et al. (2018). The rise of deep learning in drug discovery. Drug Discov Today, pii: S1359-6446(17)30359-8. doi: 10.1016/j.drudis.2018.01.039. [Epub ahead of print] [PMID:29366762]
Big Data in Drug Discovery
(1) Brown N et al. (2018). Big Data in Drug Discovery. Progress in Medicinal Chemistry, doi:10.1016/bs.pmch.2017.12.003. [Epub ahead of print] [Abstract]
February 2018
The G Protein-Coupled Receptors deorphanization landscape
Comments by Julien Hanson, University of Liege
This paper (1) sheds some light on the current state of the field and the phenomenon of reduced discoveries in the orphan landscape. Although it is true that fewer deorphanizations have been reported recently compared to the 1990-2000 period, the authors propose that the rate has reached a "steady-state" stage. Nevertheless, with more than 100 remaining orphans, the daunting task of full deorphanization that lies ahead will require creative approaches both at the technical and conceptual level. Read the full article on our blog 
(1) Laschet C, Dupuis N, Hanson J. (2018). The G Protein-Coupled Receptors deorphanization landscape. Biochem Pharmacol., pii: S0006-2952(18)30073-X. doi: 10.1016/j.bcp.2018.02.016. [Epub ahead of print] [PMID:29454621]
Integrative omics for health and disease
(1) Karczewski KJ and Snyder MP. (2018). Integrative omics for health and disease. Nature Reviews Genetics, doi:10.1038/nrg.2018.4. [Epub ahead of print] [Full text]
Selective Photoaffinity Probe That Enables Assessment of Cannabinoid CB2 Receptor Expression and Ligand Engagement in Human Cells
(1) Soethoudt M et al. (2018). Selective Photoaffinity Probe That Enables Assessment of Cannabinoid CB2 Receptor Expression and Ligand Engagement in Human Cells. J Am Chem Soc., doi: 10.1021/jacs.7b11281. [Epub ahead of print] [PMID:29420021]
Caveat usor: assessing differences between major chemistry databases
(1) Southan C. (2018). Caveat usor: assessing differences between major chemistry databases. ChemMedChem., doi: 10.1002/cmdc.201700724. [Epub ahead of print] [PMID:29451740]
£54 million funding to transform UK health through data science
Health Data Research UK is awarding £30 million funding to six sites across the UK to address challenging healthcare issues through use of data science. Each site has world-class expertise; a track record in using health data to derive new knowledge, scientific discovery and insight; and works in close partnership with NHS bodies and the public to translate research findings into benefits for patients and populations. [Read more at Health Data Research UK]
The 100,000 Genomes Project
The project will sequence 100,000 genomes from around 70,000 people. Participants are NHS patients with a rare disease, plus their families, and patients with cancer. [Read more at Genomics England]
BACE1 inhibition more effectively suppresses initiation than progression of β-amyloid pathology
(1) Peters F et al. (2018). BACE1 inhibition more effectively suppresses initiation than progression of β-amyloid pathology. Acta Neuropathol., doi: 10.1007/s00401-017-1804-9. [Epub ahead of print] [PMID:29327084]
5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology
(1) Peng Y et al. (2018). 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology. Cell, 172(4):719-730.e14. doi: 10.1016/j.cell.2018.01.001. [PMID:29398112]
Deubiquitylating enzymes and drug discovery: emerging opportunities
(1) Harrigan JA et al. (2018). Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov., 17(1):57-78. doi: 10.1038/nrd.2017.152. [PMID:28959952]
January 2018
Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone
(1) Wang S et al. (2018). Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature, doi:10.1038/nature25758. [Epub ahead of print]. [Abstract]
The Enduring Legacy of 250 Years of Pharmacology in Edinburgh
(1) Kelly JS, Mackay AVP. (2018). The Enduring Legacy of 250 Years of Pharmacology in Edinburgh. Annu Rev Pharmacol Toxicol., 58:293-307. doi: 10.1146/annurev-pharmtox-010617-052901 [PMID:28934562]
A Serendipitous Scientist
(1) Lefkowitz RJ. (2018). A Serendipitous Scientist. Annu Rev Pharmacol Toxicol., 58:17-32. doi: 10.1146/annurev-pharmtox-010617-053149. [PMID:28715979]
Pharmacogenomics of GPCR Drug Targets
Comments by Alexander Hauser, University of Copenhagen and GPCRdb
A collaboration between the MRC Laboratory of Molecular Biology, Cambridge (UK), the Scripps Research Institute in Florida and the Department of Drug Design and Pharmacology, University of Copenhagen (home of the GPCRdb team) has now published a new detailed study on the effects of genetic variation in G protein-coupled receptors on responses to FDA-approved drugs [1]. The authors address the following main questions: How variable are GPCR drug targets in the human population? Are individuals with variant receptors likely to respond differently to drugs? What is the estimated economic burden associated with variation in GPCR drug targets? Read the full article on our blog 
(1) Hauser AS et al. (2017). Pharmacogenomics of GPCR Drug Targets. Cell, 172(1-2):41-54.e19. doi:10.1016/j.cell.2017.11.033. [PMID:29249361]
Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor
(1) Che T et al. (2018). Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor. Drug Discov Today, 172(1-2):55-67.e15. doi: 10.1016/j.cell.2017.12.011. [PMID:29307491]
Drug target residence time: a misleading concept
(1) Folmer RHA. (2017). Drug target residence time: a misleading concept. Drug Discov Today, S1359-6446(17)30241-6. doi: 10.1016/j.drudis.2017.07.016. [PMID:28782685]
GPCRs as targets for approved drugs: How many targets and how many drugs?
(1) Sriram K & Insel PA. (2018). GPCRs as targets for approved drugs: How many targets and how many drugs? Mol Pharmacol., doi: 10.1124/mol.117.111062. [Epub ahead of print]. [PMID:29298813]
International Union of Basic and Clinical Pharmacology CIII
(1) Kennedy AJ & Davenport AP. (2018). International Union of Basic and Clinical Pharmacology CIII: Chemerin Receptors CMKLR1 (Chemerin1) and GPR1 (Chemerin2) Nomenclature, Pharmacology, and Function. Pharmacol Rev., 70(1):174-196. doi: 10.1124/pr.116.013177. [PMID:29279348]
Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727
(1) Roberston N et al. (2018). Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727. Nature, 553(7686):111-114 doi: 10.1038/nature25025. [PMID:29300009]
Structure of the glucagon receptor in complex with a glucagon analogue
(1) Zhang H et al. (2018). Structure of the glucagon receptor in complex with a glucagon analogue. Nature, 553(7686):106-110. doi: 10.1038/nature25153. [PMID:29300013]
Allosteric Coupling of Drug Binding and Intracellular Signaling in the A2A Adenosine Receptor
(1) Eddy MT et al. (2017). Allosteric Coupling of Drug Binding and Intracellular Signaling in the A2A Adenosine Receptor. Cell Chem Biol., 172(1-2):68-80.e12. doi: 10.1016/j.cell.2017.12.004. [PMID:29290469]
Drug Target Commons: A Community Effort to Build a Consensus Knowledge Base for Drug-Target Interactions
(1) Tang J et al. (2017). Drug Target Commons: A Community Effort to Build a Consensus Knowledge Base for Drug-Target Interactions. Cell Chem Biol., S2451-9456(17)30426-9. doi: 10.1016/j.chembiol.2017.11.009. [PMID:29276046]
A dynamic map for learning, communicating, navigating and improving therapeutic development
(1) Wagner J et al. (2017). A dynamic map for learning, communicating, navigating and improving therapeutic development. Nat Rev Drug Discov., [EPub ahead of print]. doi: 10.1038/nrd.2017.217. [PMID:29269942]
Residue-Specific Peptide Modification: A Chemist's Guide
(1) deGruyter JN et al. (2017). Residue-Specific Peptide Modification: A Chemist's Guide. Biochemistry., 56(30):3863-3873. doi: 10.1021/acs.biochem.7b00536. [PMID:28653834]
Phenome-wide association studies (PheWAS) across large "real-world data" population cohorts support drug target validation
(1) Diogo D et al. (2017). Phenome-wide association studies (PheWAS) across large "real-world data" population cohorts support drug target validation BioRxiv., doi: https://doi.org/10.1101/218875. [Full text]
The spectrum of T cell metabolism in health and disease.
(1) Bantug GR et al. (2017). The spectrum of T cell metabolism in health and disease. Nat Rev Immunol., 18(1):19-34. doi: 10.1038/nri.2017.99. [PMID:28944771]
Genetic variation in human drug-related genes
(1) Schärfe CPI et al. (2017). Genetic variation in human drug-related genes. Genome Med., 9(1):117. doi: 10.1186/s13073-017-0502-5. [PMID:29273096]
Complex Portal - A Unifying Protein Complex Database
(1) Meldal, B et al. (2017). Complex Portal - A Unifying Protein Complex Database. Genomics and Computational Biology, [S.l.], v. 4, n. 1, p. e100052, dec. 2017. ISSN 2365-7154. [Full text]
2017
December 2017
Trends in GPCR drug discovery: new agents, targets and indications
Comments by Alexander Hauser and David Gloriam, University of Copenhagen and GPCRdb
New avenues for GPCR drug discovery have emerged owing to recent advances in receptor pharmacology, technological breakthroughs in structural biology and innovations in biotechnology. A collaboration between the Department of Drug Design and Pharmacology, University of Copenhagen (home of the GPCRdb team) and the Uppsala University have published a detailed analysis of all GPCR drugs and agents in clinical trials, which reveals current trends across molecule types, drug targets and therapeutic indications [1]. Read the full article on our blog 
Hauser AS et al. (2017). Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov., 16(12):829-842. doi: 10.1038/nrd.2017.178. [PMID:29075003]
Noncoding RNAs: Master Regulators of Inflammatory Signaling
(1) Chew CL et al. (2017). Noncoding RNAs: Master Regulators of Inflammatory Signaling. Trends in Molecular Medicine, doi:10.1016/j.molmed.2017.11.003. [Epub ahead of print] [Full text]
Refining The Accuracy Of Validated Target Identification Through Coding Variant Fine-Mapping In Type 2 Diabetes
(1) Mahajan A et al. (2017). Refining The Accuracy Of Validated Target Identification Through Coding Variant Fine-Mapping In Type 2 Diabetes. bioRxiv, doi: 10.1101/144410. [Epub ahead of print] [Full text]
Genetic architecture: the shape of the genetic contribution to human traits and disease
(1) Timpson NJ et al. (2017). Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat Rev Genet., doi: 10.1038/nrg.2017.101. [Epub ahead of print] [29225335]
G Protein-Coupled Receptors Targeting Insulin Resistance, Obesity, and Type 2 Diabetes Mellitus.
(1) Riddy DM et al. (2017). G Protein-Coupled Receptors Targeting Insulin Resistance, Obesity, and Type 2 Diabetes Mellitus. Pharmacol Rev., 70(1):39-67. doi: 10.1124/pr.117.014373. [29233848]
The rise and fall of a scaffold: A trend analysis of scaffolds in the medicinal chemistry literature
(1) Zdrazil B, Guha R. (2017). The rise and fall of a scaffold: A trend analysis of scaffolds in the medicinal chemistry literature. J Med Chem, doi: 10.1021/acs.jmedchem.7b00954. [Epub ahead of print] [29235859]
Objective, Quantitative, Data-Driven Assessment of Chemical Probes
(1) Antolin AA et al. (2017). Objective, Quantitative, Data-Driven Assessment of Chemical Probes. Cell Chemical Biology, doi:10.1016/j.chembiol.2017.11.004. [Epub ahead of print] [Full text]
A snapshot of 3649 Web-based services published between 1994 and 2017 shows a decrease in availability after 2 years
(1) Osz Á et al. (2017). A snapshot of 3649 Web-based services published between 1994 and 2017 shows a decrease in availability after 2 years. Brief Bioinform., doi: 10.1093/bib/bbx159. [Epub ahead of print] [29228189]
Science Forum: The Human Cell Atlas
(1) Regev A et al. (2017). Science Forum: The Human Cell Atlas. Elife, 6. pii: e27041. doi: 10.7554/eLife.27041. [Epub ahead of print] [29206104]
Matching disease and phenotype ontologies in the ontology alignment evaluation initiative
(1) Harrow I et al. (2017). Matching disease and phenotype ontologies in the ontology alignment evaluation initiative. J Biomed Semantics., 8(1):55. doi: 10.1186/s13326-017-0162-9. [PMID:29197409]
Structural Mapping of Adenosine Receptor Mutations: Ligand Binding and Signaling Mechanisms
(1) Jespers W et al. (2017). Structural Mapping of Adenosine Receptor Mutations: Ligand Binding and Signaling Mechanisms. Trends Pharmacol Sci., pii: S0165-6147(17)30218-3. doi: 10.1016/j.tips.2017.11.001. [Epub ahead of print] [PMID:29203139]
Pharma's broken business model: An industry on the brink of terminal decline
(1) Stott K. (2017). Pharma's broken business model: An industry on the brink of terminal decline. [Blog]
The Promise & Peril in YOUR Genes
(1) Green R. (2017). The Promise & Peril in YOUR Genes. [Podcast]
Structural coverage of the proteome for pharmaceutical applications
(1) Somody JC et al. (2017). Structural coverage of the proteome for pharmaceutical applications. Drug Discov Today, 22(12):1792-1799. doi: 10.1016/j.drudis.2017.08.004. [PMID:28843631]
The target landscape of clinical kinase drugs
(1) Klaeger S et al. (2017). The target landscape of clinical kinase drugs. Science, 358(6367). pii: eaan4368. doi: 10.1126/science.aan4368. [PMID:29191878]
Post-transcriptional regulation across human tissues
(1) Franks A et al. (2017). Post-transcriptional regulation across human tissues. PLoS Comput Biol., 13(5):e1005535. doi: 10.1371/journal.pcbi.1005535. [PMID:28481885]
Mechanistic enzymology in drug discovery: a fresh perspective
(1) Holdgate GA et al. (2017). Mechanistic enzymology in drug discovery: a fresh perspective. Nat Rev Drug Discov., doi: 10.1038/nrd.2017.219. [Epub ahead of print] [PMID:29192286]
A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles
(1) Subramanian A et al. (2017). A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell., 171(6):1437-1452.e17. doi: 10.1016/j.cell.2017.10.049. [PMID:29195078]
Five ways to fix statistics
(1) Leek J et al. (2017). Five ways to fix statistics. Nature., 9551(7682):557-559. doi: 10.1038/d41586-017-07522-z. [PMID:29189798]
The CompTox Chemistry Dashboard: a community data resource for environmental chemistry
(1) Williams AJ et al. (2017). The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J Cheminform., 9(1):61. doi: 10.1186/s13321-017-0247-6. [PMID:29185060]
November 2017
Boehringer Ingelheim shares well-characterized pre-clinical molecules with the scientific community at opnMe.com
(1) https://www.opnme.com/molecules
Visualizing the GPCR Network: Classification and Evolution
(1) Hu GM et al. (2017). Visualizing the GPCR Network: Classification and Evolution. Sci Rep., 7(1):15495. [PMID:29138525]
PD-L1 genetic overexpression or pharmacological restoration in hematopoietic stem and progenitor cells reverses autoimmune diabetes
(1) Ben Nasr M et al. (2017). PD-L1 genetic overexpression or pharmacological restoration in hematopoietic stem and progenitor cells reverses autoimmune diabetes. Sci Transl Med., 9(416). pii: eaam7543. [PMID:29141886]
A simple proposal for the publication of journal citation distributions
(1) Lariviere V et al. (2016). A simple proposal for the publication of journal citation distributions. bioRxiv, doi: https://doi.org/10.1101/062109. [Epub ahead of print]. [Abstract]
Improving virtual screening of G protein-coupled receptors via ligand-directed modeling
(1) Coudrat T et al. (2017). Improving virtual screening of G protein-coupled receptors via ligand-directed modeling. PLoS Comput Biol., 13(11):e1005819. doi: 10.1371/journal.pcbi.1005819. [Epub ahead of print]. [PMID:29131821]
GPCRdb in 2018: adding GPCR structure models and ligands
(1) Pándy-Szekeres G et al. (2017). GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res., doi: 10.1093/nar/gkx1109. [Epub ahead of print]. [PMID:29155946]
Gαi is required for carvedilol-induced β1 adrenergic receptor β-arrestin biased signaling
(1) Wang J et al. (2017). Gαi is required for carvedilol-induced β1 adrenergic receptor β-arrestin biased signaling. Nat Commun., 8(1):1706. [PMID:29167435]
Cryo-EM structures of Mucolipin TRP Channels in the Lysosome: Five Together at Once
Comments by Haoxing Xu, NC-IUPHAR subcommittee Chair of the Transient Receptor Potential Channels and Professor, the University of Michigan
Five independent studies, led by Youxing Jiang, Xiaochun Li, Soek-yong Lee, Maojun Yang, and Jian Yang, respectively, report a total of three TRPML1 and two TRPML3 Cryo-EM structures, all at atomic resolution, and in both closed and agonist-bound open conformations [1-5]. Read the full article on our blog 
- Zhou X et al. (2017). Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states. Nat Struct Mol Biol, doi: 10.1038/nsmb.3502. [Epub ahead of print] [PMID:9106414]
- Zhang S et al. (2017). Cryo-EM structures of the mammalian endo-lysosomal TRPML1 channel elucidate the combined regulation mechanism. Protein Cell, 8(11):834-847 [PMID:28936784]
- Schmiege P et al. (2017). Human TRPML1 channel structures in open and closed conformations. Nature, 550(7676):366-370. [PMID:29019983]
- Hirschi M et al. (2017). Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3. Nature, 550(7676):411-414 [PMID:29019979]
- Chen Q et al. (2017). Structure of mammalian endolysosomal TRPML1 channel in nanodiscs. Nature, 550(7676):415-418. [PMID:29019981]
RNA as a small molecule druggable target
(1) Rizvi NF and Smith GF. (2017). RNA as a small molecule druggable target. Bioorg Med Chem lett., 27(23):5083-5088 [PMID:29097169]
Assay interference and off-target liabilities of reported histone acetyltransferase inhibitors
(1) Dahlin JL et al. (2017). Assay interference and off-target liabilities of reported histone acetyltransferase inhibitors. Nat Commun., 8(1):1527 [PMID:29142305]
Region and cell-type resolved quantitative proteomic map of the human heart
(1) Doll S et al. (2017). Region and cell-type resolved quantitative proteomic map of the human heart. Nat Commun., 8(1):1469 [PMID:29133944]
Architecture of the human interactome defines protein communities and disease networks
(1) Huttlin EL et al. (2017). Architecture of the human interactome defines protein communities and disease networks. Nature, 545(7655):505-509 [PMID:28514442]
Drug target ontology to classify and integrate drug discovery data
(1) Lin Y et al. (2017). Drug target ontology to classify and integrate drug discovery data. J Biomed Semantics, 8(1):50 [PMID:29122012]
Extending the Structural View of Class B GPCRs
(1) de Graaf C et al. (2017). Extending the Structural View of Class B GPCRs. Trends in Biomedical Sciences, S0968-0004(17)30188-3. doi: 10.1016/j.tibs.2017.10.003. [Epub ahead of print] [PMID:29132948]
Structure-Based Design and Discovery of New M2 Receptor Agonists
(1) Fish I et al. (2017). Structure-Based Design and Discovery of New M2 Receptor Agonists. J Med Chem, doi: 10.1021/acs.jmedchem.7b01113. [Epub ahead of print] [PMID:29094937]
The structural basis of ryanodine receptor ion channel function
(1) Meissner G. (2017). The structural basis of ryanodine receptor ion channel function. J. Gen. Physiol., doi: 10.1085/jgp.201711878. [Epub ahead of print] [PMID:29122978]
Drug discovery effectiveness from the standpoint of therapeutic mechanisms and indications
(1) Shih HP et al. (2017). Drug discovery effectiveness from the standpoint of therapeutic mechanisms and indications. Nat Rev Drug Discov., doi: 10.1038/nrd.2017.194. [Epub ahead of print] [PMID:29075002]
October 2017
Directing evolution: the next revolution in drug discovery?
(1) Davis AM et al. (2017). Directing evolution: the next revolution in drug discovery? Nat Rev Drug Discov., 16(10):681-698. doi: 10.1038/nrd.2017.146 [PMID:28935911]
Drug discovery effectiveness from the standpoint of therapeutic mechanisms and indications
(1) Shih HP et al. (2017). Drug discovery effectiveness from the standpoint of therapeutic mechanisms and indications. Nat Rev Drug Discov., 7(1):13741. doi: 10.1038/nrd.2017.194. [Epub ahead of print] [PMID:29075002]
A single extracellular amino acid in Free Fatty Acid Receptor 2 defines antagonist species selectivity and G protein selection bias
(1) Sergeev E et al. (2017). A single extracellular amino acid in Free Fatty Acid Receptor 2 defines antagonist species selectivity and G protein selection bias. Sci Rep., 7(1):13741. doi: 10.1038/s41598-017-14096-3. [PMID:29061999]
Imagining the "open" university: Sharing scholarship to improve research and education
(1) McKiernan EC. (2017). Imagining the "open" university: Sharing scholarship to improve research and education. PLoS Biol., 15(10):e1002614. doi: 10.1371/journal.pbio.1002614. [PMID:29065148]
Human gene essentiality
(1) Bartha I et al. (2017). Human gene essentiality. Nature Reviews Genetics, doi:10.1038/nrg.2017.75 [Epub ahead of print] [Full text]
D4 dopamine receptor high-resolution structures enable the discovery of selective agonists
(1) Wang S et al. (2017). D4 dopamine receptor high-resolution structures enable the discovery of selective agonists. Science, 358(6361):381-386. doi: 10.1126/science.aan5468. [PMID:29051383]
Discovery of new GPCR ligands to illuminate new biology
(1) Roth BL et al. (2017). Discovery of new GPCR ligands to illuminate new biology. Nat. Chem. Biol., 13(11):1143-1151. doi: 10.1038/nchembio.2490. [PMID:29045379]
Human genomics: Cracking the regulatory code
(1) Ward MC & Gilad Y. (2017). Human genomics: Cracking the regulatory code. Nature. News & Views, 550:190-191, doi:10.1038/550190a. [Nature: News & views]
Structural Basis for G Protein-Coupled Receptor Activation
(1) Maglik A & Kruse AC. (2017). Structural Basis for G Protein-Coupled Receptor Activation. Biochemistry, doi: 10.1021/acs.biochem.7b00747. [Epub ahead of print] [PMID:28967738]
The Human Protein Atlas - A spatial map of the human proteome
(1) Thul PJ & Lindskog C. (2017). The Human Protein Atlas - A spatial map of the human proteome. Protein Sci., doi: 10.1002/pro.3307. [Epub ahead of print] [PMID:28940711]
Functional characterization of 3D-protein structures informed by human genetic diversity
(1) Hicks M et al. (2017). Functional characterization of 3D-protein structures informed by human genetic diversity.bioRxiv., https://doi.org/10.1101/182287. [bioRxiv: Abstract]
Obesity: Receptors identified for a weight regulator
(1) Saarma M & Goldman A. (2017). Obesity: Receptors identified for a weight regulator. Nature. News & Views, doi:10.1038/nature24143. [Nature: News & views]
The drug-maker's guide to the galaxy
(1) Mullard A. (2017). The drug-maker's guide to the galaxy. Nature, 549(7673):445-447 doi:10.1038/549445a. [PMID: News Feature]
September 2017
CHEMGENIE: integration of chemogenomics data for applications in chemical biology
(1) Kutchukian PS et al. (2017). CHEMGENIE: integration of chemogenomics data for applications in chemical biology. Drug Discov Today., pii: S1359-6446(17)30102-2. doi: 10.1016/j.drudis.2017.09.004. [Epub ahead of print] [PMID:28917822]
A new research avenue investigating mitochondrial GPCR biology
Comments by Michael Spedding, Secretary General, IUPHAR, and CEO, Spedding Research Solutions SARL, France
As one of the first propositions for GPCRs being present in mitochondrial membranes, a recent report from Robert Friedlander and colleagues [1] follows on from previous work characterising synaptic and extrasynaptic mitochondria in human cortex (post-mortem samples) and their role in neuroprotection. This work, if reproduced, opens up new vistas, and has many implications for neurodegenerative diseases. Taken together, Suofu et al. show that melatonin is synthesised in mitochondria, that MT1 receptors are present in mitochondrial membranes, and that MT1 receptor stimulation reduces cytochrome c and caspase secretion caused by calcium overload. The authors propose that this is a mechanism for the neuroprotective effects of melatonin in hypoxic-ischaemic brain injury in neonatal and in models of Huntington’s disease, where there is mitochondrial impairment. Read the article on our blog 
(1) Suofu Y et al. (2017). Dual role of mitochondria in producing melatonin and driving GPCR signaling to block cytochrome c release. Proc Natl Acad Sci U S A., 114(38):E7997-E8006. doi: 10.1073/pnas.1705768114. [PMID:28874589]
Receptor Quaternary Organization Explains G Protein-Coupled Receptor Family Structure
(1) Felce JH et al. (2017). Receptor Quaternary Organization Explains G Protein-Coupled Receptor Family Structure. Cell Rep., 20(11):2654-2665. [PMID:28903045]
Understanding enzyme function evolution from a computational perspective
(1) Tyzack JD et al. (2017). Understanding enzyme function evolution from a computational perspective. Curr Opin Struct Biol., 47:131-139. [PMID:28892668]
Protein maps chart the causes of disease
(1) Fessenden M. (2017). Protein maps chart the causes of disease. Nature, 549(7671):293-295. [PMID:28905898]
Is systems pharmacology ready to impact upon therapy development? A study on the cholesterol biosynthesis pathway
(1) Benson H et al. (2017). Is systems pharmacology ready to impact upon therapy development? A study on the cholesterol biosynthesis pathway. Br J Pharmacol., doi: 10.1111/bph.14037. [Epub ahead of print] [PMID:28910500]
A natural ligand for the orphan receptor GPR15 modulates lymphocyte recruitment to epithelia
(1) Suply T et al. (2017). A natural ligand for the orphan receptor GPR15 modulates lymphocyte recruitment to epithelia. Sci Signal., 10(496). pii: eaal0180. [PMID:28900043]
Structural and Functional View of Polypharmacology
(1) Moya-García A et al. (2017). Structural and Functional View of Polypharmacology. Sci Rep., 7(1):10102. [PMID:28860623]
The T cell antigen receptor: the Swiss army knife of the immune system
(1) Attaf M et al. (2017). The T cell antigen receptor: the Swiss army knife of the immune system. Clin Exp Immunol., 181(1):1-18. [PMID:25753381]
Crystal structure of LPA6, a receptor for lysophosphatidic acid, at 3.2A
Comments by Jerold Chun (Neuroscience Drug Discovery, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, USA)
Lysophospholipids (LPs) have myriad roles as extracellular signals that activate cognate G protein-coupled receptors (GPCRs) (2). LPs for which receptors have been reported include lysophosphatidic acid (LPA) (receptors: LPA1-6), sphingosine 1-phosphate (S1P1-5), lysophosphatidyl serine (LPS1-3, 2L (2L is a pseudogene in humans)) and lysophosphatidyl inositol/glucose (LPI/LPG), all of which are Class A GPCRs. Of these 15 LP receptors, crystal structures of two have been previously reported for S1P1 (2.8-3.35A) (3) and LPA1 (2.9-3.0A) (4) both of which utilized human cDNA sequences bound in the presence of antagonists. The new structure (1), from the laboratories of Junken Aoki and Osamu Nureki, elucidates a zebrafish receptor – with 80% amino acid similarity to human LPA6, in the transmembrane (TM) region - in the absence of a ligand, which nonetheless crystalized. This contrasts with the prior 2 antagonist-bound human structures. All 3 receptors were chimeric proteins stabilized by T4-lysozyme (S1P1 and LPA6) or thermostabilized apocytochrome b562RIL (LPA1) fused to the 3rd intracellular loop, but all were capable of responding to native ligands. Read the full article on our blog 
(1) Taniguchi R et al. (2017). Structural insights into ligand recognition by the lysophosphatidic acid receptor LPA6. Nature, 548: 356-360. [PMID:28792932]
(2) Kihara Y et al. (2014) Lysophospholipid receptor nomenclature review: IUPHAR Review 8. Br J Pharmacol, 171: 3575-3594. [PMID:24602016]
(3) Hanson MA et al. (2012) Crystal structure of a lipid G protein-coupled receptor. Science, 335: 851-855. [PMID:22344443]
(4) Chrencik JE et al. (2015) Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1. Cell, 161: 1633-1643. [PMID:26091040]
Comparing structural and transcriptional drug networks reveals signatures of drug activity and toxicity in transcriptional responses
(1) Sirci F et al. (2017). Comparing structural and transcriptional drug networks reveals signatures of drug activity and toxicity in transcriptional responses. NPJ Syst Biol Appl., 3:23. [PMID:28861278]
Commensal bacteria make GPCR ligands that mimic human signalling molecules
(1) Cohen LJ et al. (2017). Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature, doi: 10.1038/nature23874. [Epub ahead of print] [PMID:28854168]
In silico prediction of novel therapeutic targets using gene–disease association data
(1) Ferrero E et al. (2017). In silico prediction of novel therapeutic targets using gene–disease association data. J Transl Med., 15(1):182. [PMID:28851378]
An atlas of B-cell clonal distribution in the human body
(1) Meng W et al. (2017). An atlas of B-cell clonal distribution in the human body. Nat Biotechnol., doi: 10.1038/nbt.3942. [Epub ahead of print] [PMID:28829438]
Functional characterization of 3D-protein structures informed by human genetic diversity
(1) Hicks M et al. (2017). Functional characterization of 3D-protein structures informed by human genetic diversity. bioRxiv, doi:10.1101/182287. [Epub ahead of print] [Abstract]
GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and nonhuman primates
(1) Mullican SE et al. (2017). GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and nonhuman primates. Nat Med., doi: 10.1038/nm.4392. [Epub ahead of print] [PMID:28846097]
Applying Structure-Based Drug Design Approaches to Allosteric Modulators of GPCRs.
(1) Congreve M et al. (2017). Applying Structure-Based Drug Design Approaches to Allosteric Modulators of GPCRs. Trends Pharmacol Sci., 38(9):837-847. [PMID:28648526]
Mechanism of intracellular allosteric β2AR antagonist revealed by X-ray crystal structure
(1) Liu X et al. (2017). Mechanism of intracellular allosteric β2AR antagonist revealed by X-ray crystal structure. Nature, 548(7668):480-484 [PMID:28813418]
Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease
(1) Ridker PM et al. (2017). Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N Engl J Med., doi: 10.1056/NEJMoa1707914. [Epub ahead of print] [PMID:28845751]
Pepcan-12 (RVD-hemopressin) is a CB2 receptor positive allosteric modulator constitutively secreted by adrenals and in liver upon tissue damage.
(1) Petrucci V et al. (2017). Pepcan-12 (RVD-hemopressin) is a CB2 receptor positive allosteric modulator constitutively secreted by adrenals and in liver upon tissue damage. Sci Rep., 25;7(1):9560. [PMID:28842619]
August 2017
FZD6 dimers dissociate after stimulation – briefly
Comments by Nevin A. Lambert (Department of Pharmacology and Toxicology Medical College of Georgia, Augusta University, USA)
GPCRs of all classes are widely thought to form homodimers, heterodimers and higher-order oligomers. The functional significance of dimerization is well understood for Class C receptors but less certain for the other GPCR classes, including the rather unconventional class F or Frizzled (FZD) receptors. Although the relationship between receptor activity and quaternary structure is often unclear, across classes it is generally found that ligand binding does not dramatically influence dimerization. A recent report by Gunnar Schulte and his colleagues suggests that in this respect class F receptors may once again be somewhat different [1]. Using an impressive combination of live-cell imaging, biochemical and modeling techniques the group presents evidence that FZD6 forms relatively stable dimers that dissociate when stimulated with the activating ligand WNT-5A. Remarkably, FZD6 protomers reassociate at the cell surface after 20 minutes of continuous stimulation, a timing which coincides with termination of ERK1/2 phosphorylation. Read the full article on our blog 
(1) Petersen J et al. (2017) Agonist-induced dimer dissociation as a macromolecular step in G protein-coupled receptor signaling. Nat Commun., 8(1):226. [PMID:28790300]
FTBMT, a novel and selective GPR52 agonist, demonstrates antipsychotic-like and procognitive effects in rodents revealing a potential therapeutic agent for schizophrenia.
(1) Nishiyama K et al. (2017). FTBMT, a novel and selective GPR52 agonist, demonstrates antipsychotic-like and procognitive effects in rodents revealing a potential therapeutic agent for schizophrenia. Journal of Pharmacology and Experimental Therapeutics, DOI: 10.1124/jpet.117.242925 [Epub ahead of print] [Abstract]
Anti-inflammatory therapies for cardiovascular disease
(1) Ridker PM and Lüscher TF. (2014). Anti-inflammatory therapies for cardiovascular disease. Eur Heart J., 35(27):1782-91. [PMID:24864079]
A Review of Recent Advances in Translational Bioinformatics: Bridges from Biology to Medicine
(1) Vamathevan J and Birney E. (2017). A Review of Recent Advances in Translational Bioinformatics: Bridges from Biology to Medicine. Yearbook of Medical Informatics, 26(1):178-187. [Abstract]
The 10,000 Immunomes Project: A resource for human immunology
(1) Zalocusky KA et al. (2017). The 10,000 Immunomes Project: A resource for human immunology. bioRxiv., doi: https://doi.org/10.1101/180489. [bioRxiv:180489]
Cannabinoid Receptor-Related Orphan G Protein-Coupled Receptors
(1) Irving A et al. (2017). Cannabinoid Receptor-Related Orphan G Protein-Coupled Receptors. Adv. Pharmacol., 80:223-247. [PMID:28826536]
Capturing LTA4 hydrolase in action: Insights to the chemistry and dynamics of chemotactic LTB4 synthesis
(1) Stsiapanava A et al. (2017). Capturing LTA4 hydrolase in action: Insights to the chemistry and dynamics of chemotactic LTB4 synthesis. PNAS USA., 114(36):9689-9604. [PMID:28827365]
Settling the score: variant prioritization and Mendelian disease
(1) Ellbeck K et al. (2017). Settling the score: variant prioritization and Mendelian disease. Nat Rev. Genetics., DOI: 10.1038/nrg.2017.52 [Epub ahead of print]. [PMID:28804138]
A kinetic view of GPCR allostery and biased agonism
(1) Lane R et al. (2017). A kinetic view of GPCR allostery and biased agonism. Nat Chem Biol., 13(9):929-937. [PMID:28820879]
A pathology atlas of the human cancer transcriptome
(1) Uhlen M et al. (2017). A pathology atlas of the human cancer transcriptome. Science., d357(6352). pii: eaan2507. [PMID:28818916]
X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog
(1) Shihoya W et al. (2017). X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog. Nat Struct Mol Biol., doi: 10.1038/nsmb.3450. [Epub ahead of print] [PMID:28805809]
The immunopathology of sepsis and potential therapeutic targets
(1) van der Poll T et al. (2017). The immunopathology of sepsis and potential therapeutic targets. Nat Rev Immunol., 17(7):407-420. [PMID:28436424]
The PI3K Pathway in Human Disease
(1) Fruman DA et al. (2017). The PI3K Pathway in Human Disease. Cell, 170(4):605-635. [PMID:28802037]
Diabetic nephropathy - is this an immune disorder?
(1) Tesch GH. (2017). Diabetic nephropathy - is this an immune disorder? Clin Sci (Lond)., 131(16):2183-2199. [PMID:28760771]
Psychosis: an autoimmune disease?
(1) Al-Diwani AAJ et al. (2017). Psychosis: an autoimmune disease? Immunology, doi:10.1111/imm.12795. [Epub ahead of print] [28704576]
Identification of essential genes for cancer immunotherapy
(1) Patel SJ et al. (2017). Identification of essential genes for cancer immunotherapy. Nature, doi: 10.1038/nature23477. [Epub ahead of print] [PMID:28783722]
Worldwide Distribution of Cytochrome P450 Alleles: A Meta-analysis of Population-scale Sequencing Projects
(1) Zhou Y et al. (2017). Worldwide Distribution of Cytochrome P450 Alleles: A Meta-analysis of Population-scale Sequencing Projects. Clin Pharmacol Ther., doi: 10.1002/cpt.690. [Epub ahead of print] [PMID:28378927]
The new alchemy: Online networking, data sharing and research activity distribution tools for scientists
(1) Williams AJ et al. (2017). The new alchemy: Online networking, data sharing and research activity distribution tools for scientists. F1000Research, 6:1315. [Abstract]
In Silico Absorption, Distribution, Metabolism, Excretion, and Pharmacokinetics (ADME-PK): Utility and Best Practices
(1) Lombardo F et al. (2017). In Silico Absorption, Distribution, Metabolism, Excretion, and Pharmacokinetics (ADME-PK): Utility and Best Practices. An Industry Perspective from the International Consortium for Innovation through Quality in Pharmaceutical Development. J Med Chem., doi: 10.1021/acs.jmedchem.7b00487. [Epub ahead of print] [PMID:28609624]
Calls grow to tap the gold mine of human genetic knockouts
(1) Mullard A. (2017). Calls grow to tap the gold mine of human genetic knockouts. Nat Rev Drug Discov., 16(8):515-518. [PMID:28757624]
A Genetic Variant Associated with Five Vascular Diseases Is a Distal Regulator of Endothelin-1 Gene Expression
(1) Gupta RM et al. (2017). A Genetic Variant Associated with Five Vascular Diseases Is a Distal Regulator of Endothelin-1 Gene Expression. Cell, 170(3):522-533.e15. [PMID:28753427]
Objective, Quantitative, Data-Driven Assessment of Chemical Probes
(1) Antolin AA et al. (2017). Objective, Quantitative, Data-Driven Assessment of Chemical Probes. bioRxiv, doi:10.1101/168369 [Epub ahead of print] [Abstract]
Choose and Use Your Chemical Probe Wisely to Explore Cancer Biology.
(1) Blagg J, Workman P. (2017). Choose and Use Your Chemical Probe Wisely to Explore Cancer Biology. Cancer Cell, 32(1):9-25. [PMID:28697345]
How Ligands Illuminate GPCR Molecular Pharmacology.
(1) Wacker D et al. (2017). How Ligands Illuminate GPCR Molecular Pharmacology. Cell, 170(3):414-427. [PMID:28753422]
Redefine Statistical Significance
(1) Benjamin D et al. (2017). Redefine Statistical Significance. PsyArXiv, doi:10.17605/OSF.IO/MKY9J. [Epub ahead of print] [Abstract]
Translating New Science Into the Drug Review Process
(1) Rouse R et al. (2017). Translating New Science Into the Drug Review Process. Therapeutic Innovation & Regulatory Science, doi:10.1177/2168479017720249. [Epub ahead of print] [Abstract]
July 2017
Channel opening and gating mechanism in AMPA-subtype glutamate receptors
(1) Twomey EC et al. (2017). Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature, doi: 10.1038/nature23479. [Epub ahead of print] [PMID:28737760]
A cryptic binding pocket in K2P2 exposes new avenues for drug development
Comments by Leigh D. Plant (Research Associate Professor, School of Pharmacy, Northeastern University)
The TREK subfamily of K2P channels (K2P2, K2P4 and K2P10) pass background potassium currents that modulate the excitability of neuronal cells and cardiac myocytes. In recent years, these channels have received significant attention as potential drug targets. In an elegant new study, Lolicato and colleagues from the Minor lab highlight the synergistic power of combining structural and functional approaches to reveal new insights into the operation of membrane proteins and unveil a new avenue for the development of TREK-channel pharmacology [1]. Read the full article on our blog 
(1) Lolicato M et al. (2017). K2P2.1 (TREK-1)-activator complexes reveal a cryptic selectivity filter binding site. Nature, 547(7663):364-368. [PMID:28693035]
Opportunities for therapeutic antibodies directed at G-protein-coupled receptors
(1) Hutchings CJ et al. (2017). Opportunities for therapeutic antibodies directed at G-protein-coupled receptors. Nat Rev Drug Discov., doi: 10.1038/nrd.2017.91. [Epub ahead of print] [PMID:28706220]
Information Retrieval and Text Mining Technologies for Chemistry
(1) Krallinger M et al. (2017). Information Retrieval and Text Mining Technologies for Chemistry. Chem Rev., 117(12):7673-7761. [PMID:28475312]
Genome-wide genetic data on ~500,000 UK Biobank participants
(1) Bycroft C et al. (2017). Genome-wide genetic data on ~500,000 UK Biobank participants. bioRxiv, 166298, doi:10.1101/166298. [Epub ahead of print] [Abstract]
Structure and function of peptide-binding G protein-coupled receptors
(1) Wu F et al. (2017). Structure and function of peptide-binding G protein-coupled receptors. J Mol Biol., doi: 10.1016/j.jmb.2017.06.022. [Epub ahead of print] [PMID:28705763]
Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity
(1) Cheng RKY et al. (2017). Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity. Cell, doi: 10.1016/j.str.2017.06.012 [Epub ahead of print] [Full text]
Phantom PAINS: Problems with the Utility of Alerts for Pan-Assay INterference CompoundS
(1) Capuzzi SJ et al. (2017). Phantom PAINS: Problems with the Utility of Alerts for Pan-Assay INterference CompoundS. J Chem Inf Model., 57(3):417-427 [PMID:28165734]
Text mining of 15 million full-text scientific articles
(1) Westergaard D et al. (2017). Text mining of 15 million full-text scientific articles. bioRxiv., doi: 10.1101/162099 [Epub ahead of print] [Abstract]
How to build a human cell atlas.
(1) Nowogrodzki A. (2017). How to build a human cell atlas. Nature, 547(7661):24-26. [PMID:28682347]
Choose and Use Your Chemical Probe Wisely to Explore Cancer Biology
(1) Blagg J, Workman P. (2017). Choose and Use Your Chemical Probe Wisely to Explore Cancer Biology. Cancer Cell, 32(1):9-25. [PMID:28697345]
Insufficient antibody validation challenges oestrogen receptor beta research
(1) Andersson S. (2017). Insufficient antibody validation challenges oestrogen receptor beta research. Nat Commun., 8:15840. [PMID:28643774]
Agonist-bound crystal structures of the CB1 cannabinoid receptor
Comments by Lahari Murali (@wavesml), Steve Alexander (@mqzspa), Steven Doughty and Abi Emtage (@AbiEmtage)
Antagonist bound crystal structures of GPCRs are useful in giving an insight into the molecular conformation of a receptor's inactive state whilst enabling the design of new drugs. However, they prove insufficient to understand the activation mechanism of the receptor and mediation of its physiological effects. This necessitates the study of agonist-bound structures. In this direction, Hua et al., (2017) [1] have recently reported two agonist-bound crystal structures of Cannabinoid Receptor 1 (CB1), one with a tetrahydrocannabinol derivative, AM11542 [PDB: 5XRA], and the other with a hexahydrocannabinol, AM841 [PDB: 5XR8]. Read the full article on our blog 
(1) Hua, T. et al. (2017). Crystal structures of agonist-bound human cannabinoid receptor CB1. Nature. doi:10.1038/nature23272. [PMID: 28678776]
Identifiers for the 21st century
Comments by Chris Southan (@cdsouthan)
While identifiers are not a traditional “hot topic” in pharmacology the subject is becoming increasingly important. One of the reasons is that for mechanistic pharmacology the community needs to define (and communicate) identifiers for the key entities of model organism species and strains, proteins, protein complexes, genes, sequences sequence variants, as well as the explicit molecular structures of chemicals, peptides and therapeutic biologicals (including antibodies) used for experimentation. Indeed one of the roles of IUPHAR (as NC-IUPHAR) is to review and recommend protein target nomenclature, in collaboration with the Human Gene Nomenclature Committee (HGNC). The paper featured here is a technical review [1] of identifier qualities and best practices that facilitate large-scale data integration. Read the full article on our blog 
(1) McMurray et al. (2017). Identifiers for the 21st century: How to design, provision, and reuse persistent identifiers to maximize utility and impact of life science data. PLoS Biol. 29;15(6). [PMID:28662064].
June 2017
Comparative transcriptomics in human and mouse
(1) Breschi A et al. (2017). Comparative transcriptomics in human and mouse. Nat Rev Genet, 18(7):425-440. [PMID:28479595]
An Expanded View of Complex Traits: From Polygenic to Omnigenic
(1) Boyle EA et al. (2017). An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell, 169(7):1177-1186. [PMID:28622505]
Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40
(1) Lu J et al. (2017). Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat Struct Mol Biol., doi: 10.1038/nsmb.3417. [Epub ahead of print] [PMID:28581512]
Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A
(1) Paulino C et al. (2017). Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A. Elife, 6: e26232. [PMID:28561733]
Activity-based protein profiling reveals off-target proteins of the FAAH inhibitor BIA 10-2474
Comments by Chris Southan (@cdsouthan)
The historical context for this commentary can be found in this blogpost. This latest report, based on activity-based profiling (ABPP), constitutes the first open biochemical investigation of BIA 10-2474 (1). The ABPP results show it inhibits several lipases that are not targeted by PF04457845, a highly selective and clinically tested FAAH inhibitor. In addition BIA 10-2474 (but not PF04457845) produced substantial alterations in lipid networks in human cortical neurons. The authors are appropriately cautious in not over-extrapolating their findings to causality of pathology recorded in the unfortunate patients (see clinical report in PMID 27806235). However, biochemical and pharmacological questions still remain. One of these is that, given the initial binding interaction is no less than three orders of magnitude lower that PF004457845, it's not entirely clear why 10-2474 was chosen as the lead. Another question is the basic kinetic parameters for purified enzymes (not just crude cell extracts in vitro) are still not available. This should include at least two methods for confirming irreversibility (e.g. IC50 vs pre-incubation or using a 10-2474 radiolabeled derivative). Word has it that a BIAL paper is in preparation so this aspect might be addressed by new results. Note also there are now two compound suppliers in PubChem offering BIA 10-2474 so more experimental reports could be expected. Read the full article on our blog 
The GtoPdb entries below have been updated with key interactions from this paper and will go live at the next release.
BIA 10-2474
PF004457845
FAAH
FAAH2
(1) van Esbroeck ACM et al. (2017). Activity-based protein profiling reveals off-target proteins of the FAAH inhibitor BIA 10-2474. Science, 356(6342): 1084-1087 [PMID:28596366]
Targeting inflammation to reduce cardiovascular disease risk: a realistic clinical prospect?
(1) Welsh P et al. (2017). Targeting inflammation to reduce cardiovascular disease risk: a realistic clinical prospect? Br J Pharmacol., doi: 10.1111/bph.13818. [Epub ahead of print] [PMID:28409825]
GDF15 is a heart‐derived hormone that regulates body growth
(1) Wang T et al. (2017). GDF15 is a heart‐derived hormone that regulates body growth. EMBO Mol Med., doi: 10.15252/emmm.201707604. [Epub ahead of print] [PMID:28572090]
GPR3 and GPR6, novel molecular targets for cannabidiol
Comments by Steve Alexander (@mqzspa)
Cannabidiol is a major metabolite from the Cannabis plant, although levels vary dependent on genetic, regional, cultivation and other factors. It lacks the psychotropic nature of THC, but has been reported to have many biological effects, to the extent that clinical trials for infantile intractable epilepsy are currently ongoing in the US. GPR3 and GPR6 are orphan GPCRs, which have previously been reported to elevate cAMP levels constitutively when expressed in recombinant systems. Although there was some evidence for activation by sphingosine 1-phosphate, this was not reproduced. In this report, a number of endogenous and Cannabis-derived metabolites were examined for their effects on β-arrestin2 recruitment in cells expressing either GPR3 or GPR6. Of these agents, only CBD caused a reduction in β-arrestin2 recruitment in a concentration-dependent manner, with pIC50 values of 5.9 and 6.7 at GPR3 and GPR6, respectively. The authors suggest that the inverse agonist nature of CBD at these receptors might be of relevance for neurodegenerative disorders, such as Parkinson’s and Alzheimer’s Disease. Read the full article on our blog 
(1) Laun AS, Song ZH. (2017) GPR3 and GPR6, novel molecular targets for cannabidiol. Biochem Biophys Res Commun. doi: 10.1016/j.bbrc.2017.05.165. [Epub ahead of print] [PMID:28571738]
Crystal structure of the GLP-1 receptor bound to a peptide agonist.
(1) Jazayeri A et al. (2017). Crystal structure of the GLP-1 receptor bound to a peptide agonist. Nature, doi: 10.1038/nature22800. [Epub ahead of print] [PMID:28562585]
Identification of the gene that codes for the σ2 receptor
(1) Alon A et al. (2017). Identification of the gene that codes for the σ2 receptor. Proc Natl Acad Sci U S A., doi: 10.1073/pnas.1705154114. [Epub ahead of print] [PMID:28559337]
May 2017
Structure of the human multidrug transporter ABCG2.
(1) Taylor NMI et al. (2017). Architecture of the human interactome defines protein communities and disease networks. Nature, 2017 May 29. doi: 10.1038/nature22345. [Epub ahead of print] [PMID:28554189].
Microbiota in T-cell homeostasis and inflammatory diseases
(1) Lee N. & Kim W-U. (2017). Microbiota in T-cell homeostasis and inflammatory diseases. Experimental & Molecular Medicine, 49, e340; doi:10.1038/emm.2017.36. [Full text].
Architecture of the human interactome defines protein communities and disease networks
(1) Huttlin E.L. et al. (2017). Architecture of the human interactome defines protein communities and disease networks. Nature, 545(7655):505-509. [PMID:28514442].
Structural Basis for Apelin Control of the Human Apelin Receptor
Comments by Anthony Davenport, David Huggins, Janet Maguire, and Robert Glen
Activation of the apelin receptor by the peptides apelin or Elabela/Toddler mediates vasodilatation and positive inotropic effects in the adult cardiovascular system and knocking out the receptors results in failure of the heart to develop in developing embryos. To date, only a limited number of Family A structures (including opioid, endothelin ETB, and orexin OX1 and OX2) have been deduced using X-ray crystallography. Ma et al., (2017) have recently reported the 2.6-Å resolution crystal structure of human apelin (APJ) receptor in complex with a synthetic 17-amino-acid apelin analogue agonist. The authors identify a two-site ligand binding mode that has not been seen in previous solved Family A receptor structures. The structure is in reasonable agreement with studies using NMR (Langelaan et al 2013) and molecular dynamics simulations (Macaluso and Glen 2010, Yang et al, 2017) In addition, many of the key interfacial receptor:agonist residues identified from the crystal structure are in agreement with mutation data on apelin binding.
Continue reading the full commentary on our blog 
(1) Ma Y et al. (2017). Structural Basis for Apelin Control of the Human Apelin Receptor. Structure, 6:858-866.e4. [PMID:28528775].
Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein
(1) Zhang Y et al. (2017). Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature, doi:10.1038/nature22394. [Epub ahead of print]. [Full text].
Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand
Comments by Shane C. Wright, Gunnar Schulte (Karolinska Institutet)
The recently published structure of a full-length SMO bound to the stabilizing compound TC114 builds on emerging concepts from earlier crystal structures of SMO and provides novel insight into how structural rearrangements of the CRD relative to the receptor core coordinate receptor activation while relating this to WNT receptors [1]. Read the full article on our blog 
(1) Zhang X et al. (2017). Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand. Nat Commun., 8:15383. [PMID:28513578].
Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators
Comments by Patrick Sexton (Monash University, Melbourne)
The glucagon-like peptide-1 receptor (GLP-1R) is a major target for treatment of Type 2 diabetes but has been refractory to the development of small molecule compounds as potential therapeutics. Song et al., (1) report the first crystal structures of the GLP-1R transmembrane domain in complex with 2 distinct negative allosteric modulators (NAMs) (PF-06372222 and NNC0640). Read the full article on our blog 
(1) Song G et al. (2017). Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature, doi: 10.1038/nature22378. [Epub ahead of print]. [PMID:28514449].
Structure of the full-length glucagon class B G-protein-coupled receptor
Comments by Laurence J. Miller (Mayo Clinic, Scottsdale, AZ, USA)
Zhang et al. (1) now report a crystal structure of full length glucagon receptor (GCGR) in an inactive conformation stabilized by the non-peptidyl antagonist, NNC0640, and mAb1, bound to the ECD. In this new structure, the ECD is elongated above the TMD, with mAb1 resting on extracellular loop 1 (ECL1), and with the stalk region that links the two dominant receptor domains present in a β-strand conformation lying across the helical bundle between ECL1 and ECL2/ECL3.
Read the full article on our blog 
(1) Zhang H et al. (2017). Structure of the full-length glucagon class B G-protein-coupled receptor. Nature, doi: 10.1038/nature22363. [Epub ahead of print]. [PMID:28514451].
Selectivity determinants of GPCR–G-protein binding
Comments by Chris Southan (@cdsouthan)
As a detailed comparative sequence/structure/evolution analysis it is relatively unusual (in a good sense) to see such a bioinformatics article in Nature. This tour de force was a collaboration between MRC Laboratory of Molecular Biology, Cambridge UK and the Department of Drug Design and Pharmacology, University of Copenhagen (home of the GPCRdb team). As we know, GPCR signal transduction involves the binding of ligand-activated receptors to their appropriate Gα proteins. In this work selectivity-determining positions for signal transduction (as structural "barcodes") were inferred by comprehensively comparing the sequence conservation between paralogues and orthologues, incorporating information from recent structures. The residue positions for the interaction interfaces are collated and presented at gpcrdb.org (tab ‘Signal Proteins’) for all human receptors and their 16 Gα proteins. This will be updated (including data from new structures) as a guide to interface determinants of coupling selectivity. Many applications of this resource can be envisaged. These could include: exploring options to target GPCR-G protein interfaces with agents that block coupling between the receptor and G protein intracellularly, protein engineering, structural studies and understanding the consequences of natural variation or rare disease associated mutations occurring in the vicinity of the barcode positions.
Note that all GtoPdb GPCRs have cross-references to GPCRdb (who we collaborate with) so users can navigate structural data (including the barcode positions) via GPCRdb, but also exploit ligand-centric navigation via GtoPdb and links out to genomic variants via the Ensembl links. Read the full article on our blog 
(1) Flock et al. (2017). Selectivity determinants of GPCR-G-protein binding. Nature, 545: 317-322. [PMID:28489817].
Protein-phospholipid interplay revealed with crystals of a calcium pump
Comments by Steve Alexander (@mqzspa)
Norimatsu and colleagues [1] have used X-ray solvent contrast modulation to assess density maps of four activation statses of SERCA1. Their observations suggest an unexpected movement of the transporter core, which allows an exaggerated ‘waving’ of the cytoplasmic-extended calcium-binding domain during the cycle of calcium transport. Read the full article on our blog 
(1) Norimatsu et al. (2017). Protein-phospholipid interplay revealed with crystals of a calcium pump. Nature, 545(7653):193-198. [PMID:28467821].
Consequences Of Natural Perturbations In The Human Plasma Proteome
(1) Sun et al. (2017). Consequences Of Natural Perturbations In The Human Plasma Proteome. bioRxiv, doi:10.1101/134551. [bioRxiv: 134551].
Assessment of the significance of patent-derived information for the early identification of compound–target interaction hypotheses
(1) Senger, S. (2017). Assessment of the significance of patent-derived information for the early identification of compound–target interaction hypotheses. Journal of Chemoinformatics, 9:26, doi: 10.1186/s13321-017-0214-2. [SpringerOpen: view article].
A High-Dimensional Atlas of Human T Cell Diversity Reveals Tissue-Specific Trafficking and Cytokine Signatures
(1) Wong et al. (2017). A High-Dimensional Atlas of Human T Cell Diversity Reveals Tissue-Specific Trafficking and Cytokine Signatures. Immunity, 45(2):442-5. [PMID: 27521270].
April 2017
New developments in cryo-electron microscopy reveal the first Class B GPCR active structure in complex with a G protein
Comments by Dr. Fiona H. Marshall (Director & CSO, Heptares Therapeutics) (@aston_fm)
Solving fully active structures in complex with G proteins is challenging. Recent advances in cryo-electron microscopy (cryo-EM) mean that near atomic resolution is becoming possible for protein complexes of smaller molecular size (sub 100kDa). Using the volta phase plate greatly increases the contrast, particularly at the lower end of the molecular size range, meaning that structures of GPCR complexes are now within reach. The first example of this has been published [1]. Read the full article on our blog 
(1) Liang et al. (2017). Phase-plate cryo-EM structure of a class B GPCR–G-protein complex. Nature, doi: 10.1038/nature22327. [PMID: 28437792].
Last rolls of the yoyo: Assessing the human canonical protein count
(1) Southan, C. (2017). Last rolls of the yoyo: Assessing the human canonical protein count. F1000Research, 6:448. [F1000: doi:10.12688/f1000research.11119.1].
Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity
(1) Saleheen et al. (2017). Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature, 544(7649):235-239. [PMID: 28406212].
The Drug Repurposing Hub: a next-generation drug library and information resource
(1) Corsello et al. (2017). The Drug Repurposing Hub: a next-generation drug library and information resource. Nat. Med., 23(4):405-408. [PMID: 28388612].
Neurokinin 3 receptor antagonism for menopausal hot flushes
Comments by Chris Southan (@cdsouthan)
Neurokinin B signalling is increased in menopausal women and has been implicated as an important mediator of hot flushes. A phase 2 trial has assessed the effectiveness of an oral neurokinin 3 receptor antagonist (MLE4901 (AZD2624)) [1]. Read the full article on our blog 
(1) Prague et al. (2017). Neurokinin 3 receptor antagonism as a novel treatment for menopausal hot flushes: a phase 2, randomised, double-blind, placebo-controlled trial. Lancet, S0140-6736(17)30823-1. [PMID: 28385352].
Fish peptide treats cardiovascular disease
Comments by Chris Southan (@cdsouthan)
The novel GPCR peptide ligand Elabela/Toddler (APELA, Ela), first identified in the fish Danio and critical for the development of the heart, has now been identified in the human cardiovascular system. Yang et al. [1] have recently showed that the peptide binds to the apelin receptor in human heart. Read the full article on our blog 
(1) Yang et al. (2017). Elabela/Toddler Is an Endogenous Agonist of the Apelin APJ Receptor in the Adult Cardiovascular System, and Exogenous Administration of the Peptide Compensates for the Downregulation of Its Expression in Pulmonary Arterial Hypertension. Circulation, 135(12):1160-1173. [PMID: 28137936].
Crystal structures of human AT2 reveal molecular mechanism for lack of desensitization and internalization
Comments by Anthony Davenport
The intracellular signal transduction processes activated by the angiotensin AT2 receptor, are atypical for a GPCR and different from the AT1 receptor. Although the classic motifs a GPCR are present in AT2 receptor; it fails to demonstrate classic features of G-protein signalling such as desensitization by phosphorylation, and receptor regulation by internalization. Zhang et al. [1] report the crystal structures of human AT2 bound to an AT2-selective ligand and to an AT1 /AT2 dual ligand, capturing the receptor in an active-like conformation. Read the full article on our blog 
(1) Zhang et al. (2017). Structural basis for selectivity and diversity in angiotensin II receptors. Nature, 544(7650):327-332. [PMID: 28379944].
The druggable genome and support for target identification and validation in drug development (3rd Apr 2017)
(1) Finan et al. (2017). The druggable genome and support for target identification and validation in drug development. Science Translational Medicine, 29(383) eaag1166. [Science Trans Med: Full text].
March 2017
FDA allows marketing of tests to provide genetic risk information
The FDA approved genetic testing company 23andMe to sell customers their susceptibility to heritable genetic traits. Many of these are pharmacologically revelevant
Structural insights into adiponectin receptors suggest ceramidase activity
Comments by Steve Alexander (@mqzspa)
Adiponectin receptors, Adipo1 and Adipo2 suggested to exhibit ceramidase activity - based on in silico evidence [1]. Read the full article on our blog 
(1) Vasiliauskaité-Brooks et al. (2017). Structural insights into adiponectin receptors suggest ceramidase activity. Nature, 544(7648):120-123. [PMID: 28329765].
Molecular Structure of the Human CFTR Ion Channel
(1) Liu et al. (2017). Molecular Structure of the Human CFTR Ion Channel. Cell, 23;169(1):85-95. [PMID: 28340353].
Bedside Back to Bench: Building Bridges between Basic and Clinical Genomic Research
(1) Manolio et al. (2017). Bedside Back to Bench: Building Bridges between Basic and Clinical Genomic Research. Cell, 169(1):6-12. [PMID: 28340351].
Drug Target Ontology to Classify and Integrate Drug Discovery Data
(1) Lin et al. (2017). Drug Target Ontology to Classify and Integrate Drug Discovery Data. bioRxiv, doi:10.1101/117564. [bioRxiv: 117564].
Functional Social network architecture of human immune cells unveiled by quantitative proteomics
(1) Rieckmann et al. (2017). Social network architecture of human immune cells unveiled by quantitative proteomics. Nat. Immunol., 18(5):583-593. [PMID: 28263321].
Functional Selectivity in Cytokine Signaling Revealed Through a Pathogenic EPO Mutation
(1) Kim et al. (2017). Functional Selectivity in Cytokine Signaling Revealed Through a Pathogenic EPO Mutation. Cell, 168(6):1053-1064. [PMID: 28283061].
An atlas of human long non-coding RNAs with accurate 5′ ends
(1) Hon et al. (2017). An atlas of human long non-coding RNAs with accurate 5′ ends. Nature, 543(7744):199-204. [PMID: 28241135].
The Ecstacy and Agony of Assay Interference Compounds
(1) Alrich et al. (2017). The Ecstacy and Agony of Assay Interference Compounds. Biochemistry, 56(10):1363-1366. [PMID: 28244742].
Structural Basis of Substrate Recognition by the Multidrug Resistance Protein MRP1
Comments by Prof. Ian Kerr (University of Nottingham) (@iankerr_science)
Despite a flurry of mammalian ATP binding cassette (ABC) transporter structures in the last 2 years the Holy Grail has still been to determine how these diverse proteins interact with their transport substrates. Jue Chen and colleagues at the Rockefeller have now accomplished this for the multidrug resistance protein-1 (MRP1/ABCC1) using advances in high resolution cryo-electron microscopy to show the structures of substrate-free and leukotriene C4 bound protein [1]. Read the full article on our blog 
(1) Johnson Z.L., Chen J. (2016). Structural Basis of Substrate Recognition by the Multidrug Resistance Protein MRP1. Cell, 9;168(6):1075-1085.e9. [PMID: 28238471].
The complete structure of an activated open sodium channel
(1) Sula et al. (2017). The complete structure of an activated open sodium channel. Nat. Commun., 8:14205. [PMID: 28205548].
Peripherally administered orexin improves survival of mice with endotoxin shock
(1) Ogawa et al. (2017). Peripherally administered orexin improves survival of mice with endotoxin shock. Elife, 30;5. [PMID: 28035899].
Targeted Elimination of G Proteins and Arrestins Defines Their Specific Contributions to Both Intensity and Duration of G Protein-coupled Receptor Signaling
(1) Alvarez-Curto et al. (2017). Targeted Elimination of G Proteins and Arrestins Defines Their Specific Contributions to Both Intensity and Duration of G Protein-coupled Receptor Signaling. J. Biol. Chem., 291(53):27147-27159. [PMID: 27852822].
Structure of the Adenosine A1 Receptor Reveals the Basis for Subtype Selectivity
(1) Glukhova et al. (2017). Structure of the Adenosine A1 Receptor Reveals the Basis for Subtype Selectivity Cell, 168(5):867-877. [PMID: 28235198].
The Orphan Receptor GPR17 Is Unresponsive to Uracil Nucleotides and Cysteinyl Leukotrienes
(1) Simon et al. (2017). The Orphan Receptor GPR17 Is Unresponsive to Uracil Nucleotides and Cysteinyl Leukotrienes. Mol. Pharmacol., 91(5):518-532. [PMID: 28254957].
Pharmacology of Modulators of Alternative Splicing
(1) Bates et al. (2017). Pharmacology of Modulators of Alternative Splicing. Pharmacol. Rev., 69(1):63-79. [PMID: 28034912].
Ligand and Target Discovery by Fragment-Based Screening in Human Cells
(1) Parker et al. (2017). Ligand and Target Discovery by Fragment-Based Screening in Human Cells. Cell, 168(3):527-541. [PMID: 28111073].
Discovery of GBT440, an Orally Bioavailable R-State Stabilizer of Sickle Cell Hemoglobin
(1) Metcalf et al. (2017). Discovery of GBT440, an Orally Bioavailable R-State Stabilizer of Sickle Cell Hemoglobin. ACS Med. Chem. Lett., 8(3):321-326. [PMID: 28337324].
Crystal Structure of an LSD-Bound Human Serotonin Receptor
(1) Wacker et al. (2017). Crystal Structure of an LSD-Bound Human Serotonin Receptor. Cell, 168(3):377-389. [PMID: 28129538].
Structures of the Human HCN1 Hyperpolarization-Activated Channel
(1) Lee C.H. & MacKinnon R. (2017). Structures of the Human HCN1 Hyperpolarization-Activated Channel. Cell, 168(1-2):111-120. [PMID: 28086084].
Structure of a Pancreatic ATP-Sensitive Potassium Channel
(1) Li et al. (2017). Structure of a Pancreatic ATP-Sensitive Potassium Channel. Cell, 168(1-2):101-110. [PMID: 28086082].
February 2017
A global genetic interaction network maps a wiring diagram of cellular function
(1) Costanzo et al. (2016). A global genetic interaction network maps a wiring diagram of cellular function. Science, 353(6306). [PMID: 27708008].
Orphan receptor ligand discovery by pickpocketing pharmacological neighbors
(1) Ngo et al. (2017). Orphan receptor ligand discovery by pickpocketing pharmacological neighbors. Nat. Chem. Biol., 13(2):235-242. [PMID: 27992882].
Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists
(1) Zheng et al. (2016). Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature, 540(7633):458-461. [PMID: 27926736].
Intracellular allosteric antagonism of the CCR9 receptor
(1) Oswal et al. (2016). Intracellular allosteric antagonism of the CCR9 receptor. Nature, 540(7633):462-465. [PMID: 27926729].
Visualizing GPCR ‘Megaplexes’ Which Enable Sustained Intracellular Signaling
(1) Marshall F. H. (2016). Visualizing GPCR ‘Megaplexes’ Which Enable Sustained Intracellular Signaling. Trends Biochem Sci, 41(12):985-986. [PMID: 27825513].
Structural basis for the gating mechanism of the type 2 ryanodine receptor RyR2
Guide to Pharmacology: Ryanodine receptor family
(1) Peng et al. (2016). Novel agonist bioisosteres and common structure-activity relationships for the orphan G protein-coupled receptor GPR139. Sci. Rep., 6, 36681. [PMID: 27830715].
January 2017
The orphan GPR139 receptor is activated by peptides
Comments by David E. Gloriam and Anne Cathrine Nøhr Jensen (Department of Drug Design and Pharmacology, University of Copenhagen)
Two new publications on the orphan class A GPCR, GPR139. The first describing the first combined structure-activity relationships of all surrogate agonists, and a common pharmacophore model for future ligand identification and optimization [1]. The second, showing that the endogenous melanocortin 4 receptor agonists,; adrenocorticotropic hormone and α- and β-melanocyte stimulating hormone in the low micromolar range. In addition, a potentially novel subpeptide (from consensus cleavage site) represents the most potent putative endogenous activator, so far (EC50 value of 600 nM) [2]. Read the full article on our blog 
(1) Shehata, M.A. (2016). Novel agonist bioisosteres and common structure-activity relationships for the orphan G protein-coupled receptor GPR139. Sci. Rep., 6, 36681. [PMID: 27830715].
(2) Nøhr, A.C. et al. (2016). The orphan G protein-coupled receptor GPR139 is activated by the peptides: Adrenocorticotropic hormone (ACTH), α-, and β-melanocyte stimulating hormone (α-MSH, and β-MSH), and the conserved core motif HFRW. Neurochem Int., 102:105-113. [PMID: 27916541].
X-ray crystallographic study defines binding domains for Ca2+ antagonist drugs and their molecular mechanism of action
Comments by Jörg Striessnig (Department of Pharmacology and Toxicology – Institute of Pharmacy, Universität Innsbruck)
This year witnessed a tremendous progress in our understanding of the structure-function relationship of voltage-gated Ca2+ channels. This is based on the cryo-electron microscopy structure of the rabbit Cav1.1 Ca2+ channel complex at a nominal resolution of 3.6 Å (see Hot Topics Sep 20, 2016) which is now nicely complemented by a study defining the binding domains for Ca2+ antagonist drugs and their molecular mechanism of action at atomic resolution [1]. Read the full article on our blog 
(1) Tang et al. (2016). Structural basis for inhibition of a voltage-gated Ca2+ channel by Ca2+ antagonist drugs. Nature, 537, 117–121. [PMID: 27556947].
2016
Will the real splice variants please stand up?
Comments by Chris Southan, view full comments on our blog (15th Nov 2016)
The number of alternative mRNA splice forms that map to human protein coding loci has increased to the point that nearly all proteins have such associated database records. This gives rise to the paradox that the gene build pipeline from the latest Ensembl GRCh38 reference genome assembly indicates 19,919 protein coding loci (which shrinks to 19,022 with HGNC annotation stringency) but 198,002 transcripts (i.e. nearly 10 transcripts per protein).
X-ray structure of the endothelin ETB receptor
Comments by Anthony Davenport, view full comments on our blog (28th Oct 2016)
Endothelin is a peptide that acts via two G-protein coupled receptors. ETA mainly causes vasoconstriction. In contrast ETB predominantly acts as a beneficial clearing receptor and by the release of endothelium derived relaxing factors, vasodilatation. This paper describes for the first time the crystal structure of the endothelin ETB receptor [1].
(1) Shihoya et al. (2016). Activation mechanism of endothelin ETB receptor by endothelin-1. Nature, 537, 363-368. [PMID: 27595334].
Synthesis and SAR for depsipeptide natural products as selective G protein inhibitors
Comments by Chris Southan, view full comments on our blog (19th Oct 2016)
A team including the Gloriam Group at the University of Copenhagen (also the home of GPCRDB) have a paper out in Nature Chemistry reporting the first total synthesis of YM-254890 and FR900359 [1] .
(1) Xiong et al. (2016). Total synthesis and structure–activity relationship studies of a series of selective G protein inhibitors. Nat. Chem., Nov;8(11):1035-1041. [PMID: 27768111].
X-ray structure of P2X3 receptor
Comments by Steve Alexander, view full comments on our blog (19th Oct 2016)
Extracellular ATP is able to activate two families of cell-surface receptors, one of which is the ligand-gated ion channel family of P2X receptors.
(1) Mansoor et al. (2016). X-ray structures define human P2X3 receptor gating cycle and antagonist action. Nature, 538:66-71. doi: 10.1038/nature19367. [PMID: 27626375].
High resolution structure of the voltage-gated skeletal muscle Ca2+ channel complex
Comments by Jörg Striessnig (Department of Pharmacology and Toxicology – Institute of Pharmacy, Universität Innsbruck)
View full comments on our blog (20th Sep 2016)
In a recent article in Nature [1], Wu et al. present the cryo-electron microscopy structure of the rabbit Cav1.1 complex at a nominal resolution of 3.6 Å.
(1) Wu et al. (2016). Structure of the voltage-gated Ca2+ channel Cav1.1 at 3.6 Å resolution. Nature, 537:191-196 [PMID: 27580036].
Allosteric Modulation of Receptor Function and Regulation
Comments by David E. Gloriam (Department of Drug Design and Pharmacology, University of Copenhagen)
View full comments on our blog (6th Sep 2016)
Changeux and Christopoulos have recently described in Cell [1] how common mechanisms link the allosteric sites of activation and response within the four major receptor families of ligand- and voltage-gated ion channels, G-protein-coupled receptors, nuclear hormone receptors, and receptor tyrosine kinases
(1) Changeux, J.-P. and A. Christopoulos (2016). Allosteric Modulation as a Unifying Mechanism for Receptor Function and Regulation. Cell, 166(5): p. 1084-1102 [PMID: 27565340].
Analysis of protein-coding genetic variation in humans
Comments by Chris Southan
View full comments on our blog
Lek et al. [1] in Nature, describes a tour de force large scale reference data set of high-quality protein-coding variation generated via the Exome Aggregation Consortium (ExAC) [2].
(1) Lek et al. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature, 536, 285–291 [PMID: 27535533].
Discovery of opioid analgesics with reduced side effects
Comments by Anthony Davenport
View full comments on our blog
Manglik et al. [1], writing in Nature, believe they may have found a new form of painkiller that works just as well as morphine but lacks its potentially lethal side effect.
(1) Manglik et al. (2016). Structure-based discovery of opioid analgesics with reduced side effects. Nature, 8;537(7619):185-190 [PMID: 27533032].
Comments by Curation Team
View full comments on our blog
The key value of our curation is the extraction of chemistry-activity-target data from papers. Giving this relationship a formal structure in our database records not only provides direct value for users but this is also propagated globally by other databases that link to and/or subsume our content...
NMDAR inhibiton-independent antidepressant actions of ketamine metabolites
Comments by Curation Team
As an allosteric modulator of the NMDA receptor ketamine has been widely clinically utilized (and abused) since its approval in 1970. However, the molecular mechanism of action (mmoa) which was never entirely clear has now been investigated in a new Nature paper (1). The authors show that metabolism of the (R, S)-ketamine racemate to the corresponding pair of 6-hydroxynorketamine (HNK) stereoisomers is essential for its antidepressant effects. They also show that, in mice, this action is largely residing in the (2R, 6R)-HNK form and is independent of NMDAR inhibition but rather involves an AMPA receptor pathway. This work thus raises the prospect of the eventual identification of a new molecular target for ketamine metabolite action. Commentary on the paper has appeared both in that issue of Nature and on the “In the Pipeline” blog. Somewhat unusually, the authors had already filed part of the data some years ago in WO2013056229 assigned to the US Government. This detailed work leads one to speculate how many other racemic mixtures in clinical use will turn out not only to have complex in vivo metabolism but either adventitious polypharmacolgy (and/or off-target liabilities) as a consequence of mmoa “splitting” between active chiral metabolites.
In addition to updating the ketamine entry given above, the following new ligands IDs have been added to the GtoPdb development server. These are 9152 for (S) ketamine, 9153 for (R) ketamine and 9154 for (2R, 6R)-HNK (these will all be available in the GtoPdb 2016.3 release (May/June 2016)). The respective PubChem entries, CID 182137, CID 644025 and CID 89504167 can provide cheminformatic links in the interim. The new Nature publication does not map the metabolite interactions with human NMDAR subunits via in vitro binding constants but if these (and for new targets) are eventually published we will add them.
(1) Zanos et al. (2016). NMDAR inhibition-independent antidepressant actions of ketamine metabolites.
Nature 4 May, Epub ahead of print. doi: 10.1038/nature17998. [PMID: 27144355]
PEN identified as ligand of orphan receptor GPR83
Comments by Anthony Davenport
PEN (amino acid sequence AADHDVGSELPPEGVLGALLRV) is a biologically active peptide generated from proSAAS along with other peptides including BigLEN, which was reported to be a ligand for the ophan receptor GPR171 (1).
GPR83 is currently classified as an orphan receptor (2) and mainly localised in the mouse to the CNS. Despite the absence of an identified endogenous ligand, GPR83 is of interest as it has been implicated in behavior, learning, and metabolic regulation. The same group have now identified PEN as a ligand activating GPR83 (3). In the ‘gold standard’ of ligand binding, N-terminally tyrosinated radioidodinated rat LEN bound with reasonable affinity ( ~8 nM).
NPY was previously reported as a possible ligand for the rat GPR83, also known as GIR (4). The present authors were unable to detect activity at 1 μM. This was in agreement with Southern et al. (5) who were also unable to detected any activity of thirteen NPY peptides and analogues tested at GPR83 linked to beta-arrestin. Comparatively high concentrations of zinc ions (0.1 mM) have also been reported to activate the receptor (6)
(1) Gomes I et al. (2013). GPR171 is a hypothalamic G protein-coupled receptor for BigLEN, a neuropeptide involved in feeding.
Proc. Natl. Acad. Sci. 110(40): 16211-6. doi: 10.1073/pnas.1312938110. [PMID: 24043826]
(2) Davenport AP et al. (2013). International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands.
Pharmacol. Rev. 65(3): 967-86. doi: 10.1124/pr.112.007179. [PMID: 23686350]
(3) Gomes I et al. (2016). Identification of GPR83 as the receptor for the neuroendocrine peptide PEN.
Sci. Signal. 9(425): ra43. doi: 10.1126/scisignal.aad0694. [PMID: 27117253]
(4) Sah R et al. (2007). Interaction of NPY compounds with the rat glucocorticoid-induced receptor (GIR) reveals similarity to the NPY-Y2 receptor.
Peptides. 28(2): 302-9. [PMID: 17240481]
(5) Southern C et al. (2013). Screening β-arrestin recruitment for the identification of natural ligands for orphan G-protein-coupled receptors.
J Biomol. Screen. 18(5): 599-609. doi: 10.1177/1087057113475480. [PMID: 23396314]
(6) Müller A et al. (2013). G-protein coupled receptor 83 (GPR83) signaling determined by constitutive and zinc(II)-induced activity.
PLoS One. 8(1): e53347. doi: 10.1371/journal.pone.0053347. [PMID: 23335960]
A crystal structure of human σ1 receptor
Comments by Steve Alexander
The sigma receptor has been a ‘receptor-in-waiting’ since extensive binding characterisation several decades ago revealed a ‘fourth’ opioid receptor distinct from the conventional GPCR, delta, kappa and mu opioid peptide receptors (and the more recently defined NOP receptor which responds to nociceptin/orphanin FQ). The sigma receptor also has functional impact, in particular in the nervous and cardiovascular systems, although with no clear molecular mechanism/s for its functional influence. The crystal structure published (1) describes a homotrimer, each subunit having a single transmembrane domain. The transmembrane domains appear to be located at the apices of a triangular structure, which is suggested by the authors to have a sufficiently hydrophobic flat perimembrane surface that it might merge with the membrane. Ligand binding is suggested to be in a deep hydrophobic pocket close to the juncture between cytosolic and transmembrane domains.
(1) Schmidt HR, Zheng S, Gurpinar E et al. (2016). Crystal structure of the human σ1 receptor.
Nature 532(7600): 527-530. doi: 10.1038/nature17391. [PMID: 27042935] [PDB: 5HK1, 5HK2]
A crystal structure of a serotonin transporter - one of the most exploited proteins therapeutically
Comments by Steve Alexander
The 5HT, or serotonin, transporter is one of the most therapeutically-exploited drug targets, through blockade by tricyclic and SSRI antidepressant drugs leading to changes in monoamine signalling in the brain. The crystal structures reported here (1) include complexes with two distinct SSRI, which appear to lock the transporter in an outward-facing conformation through binding to the 5HT binding site. An allosteric site is also suggested, which may correlate with observations of allosteric behaviour of some transport inhibitors.
(1) Coleman JA, Green EM & Gouaux E. (2016). X-ray structures and mechanism of the human serotonin transporter
Nature 532(7599): 334-339. doi: 10.1038/nature17629. [PMID: 27049939] [PDB: 5I6X, 5I6Z, 5I71, 5I73, 5I74, 5I75, 5I66]
A major step towards a therapy for acute pancreatitis
Comments by the curation team
Acute pancreatitis (AP) is a common and devastating inflammatory condition of the pancreas that can lead to systemic multiple organ dysfunction syndrome (MODS) and death. A research team that includes scientists from Edinburgh University in collaboration with GSK (https://www.ed.ac.uk/medicine-vet-medicine/news-events/latest-news/therapy-organ-failure) have recently published a paper (1) in Nature Medicine describing a major advance in the possible treatment. This includes the description of GSK180 as a potent and specific inhibitor of kynurenine-3-monooxygenase (KMO). Treatment with GSK180 resulted in rapid changes in the levels of kynurenine pathway metabolites in vivo and afforded therapeutic protection against MODS in a rat model of AP.
This not only marks a "home-grown" entry into GtoPdb but now also consititutes an entry in the pilot version of our immunopharmacology portal (link to blog).
(1) Mole DJ et al. (2016). Kynurenine-3-monooxygenase inhibition prevents multiple organ failure in rodent models of acute pancreatitis
Nat. Med. Feb; 22(2): 202-9. doi: 10.1038/nm.4020. [PMID: 26752518]
Fatty acid amide hydrolase (FAAH) inhibitors and the case of BIA 10-2474
Comments by the curation team
The clinical trial disaster of BIA 10-2474, where one of the Phase 1 trial paricipants died on the 17th of Jan 2016, has been commented on widely, both inside and outside the pharmacology community. A selection of these reports provide context and include quotes from members of NC-IUPHAR and GuidetoPHARMACOLOGY curators.
Nature, vol. 529, issue 7586: Scientist in the dark after French clinical trial proves fatal.
Science, 16 Jan 2016: More details emerge on fateful French drug trial.
BPS News, 22 Jan 2016: Improve early access to data from catastrophic clinical trials: A statement on behalf of the British Pharmacological Society.
Forbes, 18 Jan 2016: Scientists speculate on what caused the Bial drug testing tradegy in France.
Chris Southan blog: The unfortuate case of BIA-10-2474
BIA 10-2474 is a fatty acid amide hydrolase (FAAH) inhibitor that was being developed by the Portuguese company Bial (https://www.bial.com/en/r_d.2/pipeline.29/pipeline.a27.html). In humans, other primates and rabbits there are two paralogoues, FAAH and FAAH2. The amino acid sequence similarity is low and the FAAH2 ortholog is absent from rats and mice. The few studies on FAAH2 report different tissue distributions but a similar pharmacology to FAAH, with modest differences in the potency of inhibitors. Inhibition of FAAH allows accumulation of fatty acid amides, including anandamide. This activates cannabinoid receptors, the same targets as one of the active ingredients of the Cannabis plant, Δ9-tetrahydrocannabinol, THC. In the brain and spinal cord it is thought to act as a neuromodulator rather than a 'standard' neurotransmitter and may acumulate at synapses as an endogenous mechanism to reduce activity in particular pathways. The expectation is that FAAH inhibitors allow an amplification of signalling through the cannabinoid system with a range of therapeutic effects including for anxiety and motor disorders and chronic pain. Many inhibitors have been published but none of those trialed so far have shown sufficient efficacy to be progressed (but also had no adverse event indications).
The BIA 10-2474 structure was eventually released in a clinical trial protocol since there was neither a publication nor a clinical trial database entry. We have mapped this to a Bial patent but without detailed activity profiling (if any additional provenance data appears we will add this). The list of clinical inhibitors is now updated in the FAAH target entry but note that the development has of JNJ-42165279 now been halted as a precautionary measure.
Blocking a receptor on inflammatory microglial cells could protect against Alzheimer’s
Comments by the curation team
Olmos-Alonso et al. (1) generated considerable interest in the media last week. The MRC team from the University of Southampton used a transgenic mouse model of Alzheimer's-like pathology to show that inhibition of CSF1R by a tyrosine kinase inhibitor resulted in the blockade of microglial proliferation. The consequent improved memory and behavioural performance as well as prevention of synaptic degeneration, suggests the therapeutic strategy of modifying CSF1R activation could ameliorate Alzheimer's disease.
Links to the Guide to Pharmacology entries for the colony stimulating factor 1 receptor and the GW2580 ligand.
(1) Olmos-Alonso A, Schetters ST, Sri S et al. (2016). Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer's-like pathology
Brain : epub jan 8 [PMID: 26747862]
2015
What’s been hiding under the bridge? - Ogerin, an allosteric modulator of GPR68
Comments by Anthony Davenport
Huang et al. (1) in an article in Nature describe an integrated experimental and computational approach to discover ligands that they used as a probe to reveal some of the physiological functions of GPR68. This G-protein coupled receptor belongs to a proton sensing family detecting acidic pH, but to date there has been no consensus on the sellctivity and reproducibility of small molecule ligands to explore function. The authors present data on a potent GPR86 positive allosteric modulator (PAM) named ogerin, that supressed recall in fear conditioning in mice. The results implicates GPR68 in anxiety and suggests a potential new drug target in this and related CNS disorders. As proof of principle that this strategy may have wider applicability to orphan GPCRs, allosteric agonist and negative allosteric modulators were also identified for GPR68, also a member of the proton sensing family with a widespread distribution in central and peripheral tissues and is fully activated at pH 6.8, but almost silent at pH 7.8.
(1) Huang XP, Karpiak J, Kroeze WK et al. (2015). Allosteric ligands for the pharmacologically dark receptors GPR68 and GPR65
Nature 527: 477-83. [PMID: 26550826]
Loss of GPR3 reduces the amyloid plaque burden and improves memory in Alzheimer's disease mouse models.
Recommended by Michael Spedding
(1) Huang Y, Skwarek-Maruszewska A, Horré K et al. (2015). Loss of GPR3 reduces the amyloid plaque burden and improves memory in Alzheimer's disease mouse models.
Sci Transl Med. 7: 309ra164. [PMID: 26468326]
2015 Nobel Prize for drug discovery
Comments by Anthony Davenport
The Nobel Prize in Physiology or Medicine 2015 was awarded for the first time to a scientist based in China, the pharmacologist Youyou Tu for her discovery of artemisinin, identified following screening of herbal remedies for the treatment of malaria and isolated from the wormwood plant, Artemisia annua. The prize was shared with William C. Campbell and Satoshi Ōmura for their discovery of avermectin, isolated from bacterial cell cultures, that was developed as ivermectin, a novel drug effective against infections caused by roundworm parasites that lead to River Blindness (onchocerciasis) and lympharic filariasis. Ivermectin binds selectively to glutamate-gated chloride ion channels in invertebrate muscle and nerve cells, causing increased permeability of the cell membrane to chloride ions. This results in hyperpolarization of the cell, leading to paralysis and death of the roundworm. Ivermectin may also disrupt GABA-mediated central nervous system neurosynaptic transmission. Merck currently donates as Mectizan, 140 million ivermectin treatments for River Blindness and 130 million for lymphatic filariasis (co-administered with albendazole, donated by GlaxoSmithKline), annually. For more information see Nobelprize.org.
A structure for the Alzheimer's disease gamma-secretase complex
Comments by the GtoPdb team
A team from MRC Cambrige, UK has published the structure of human γ-secretase (1). The 3.4 Å resolution, cryo-electron microscopy structure (5A63) provides many insights including mechanistic explanations of mutations related to early-onset Alzheimer's disease (AD). The UniProt entry for the catalytic subunit presenilin 1 (P49768, PSEN1) lists 73 amino acid variants and the authors show these cluster at two hotspots located at the centre of a distinct four transmembrane segment bundle.
Our presenilin 1 entry lists those inhibitors that have reached clinical stages. None have proved clinically effective so far but we will add new ones as they advance. It will be of interest and importance if this breakthrough in structure determination of the complex can identify binding sites for these inhibitors and facilitate Notch-sparing optimisation.
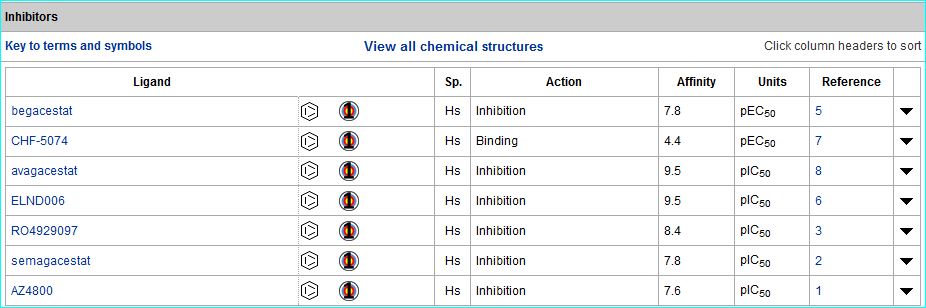
(1) Bai XC, Yan C, Yang G et al. (2015). An atomic structure of human γ-secretase.
Nature. 525: 212-7. [PMID: 26280335]
New approach to the diagnosis of atherosclerosis
Comments by the GtoPdb team
A new paper "Identifying active vascular microcalcification by (18)F-sodium fluoride positron emission tomography" represents a major step forward in the diagnosis of atherosclerosis. The report introduces vascular calcification as a hallmark of atherosclerosis. While macrocalcification confers plaque stability, microcalcification is a key feature of high-risk atheroma and is associated with increased morbidity and mortality. This study demonstrated the binding of the positron-emitting radioactive tracer, 18F sodium fluoride specifically to calcium within plaques. In terms of selectivity, specificity and pharmacodynamic parameters, this binding is similar to ligand-receptor interactions. This is the only currently available clinical imaging platform that can non-invasively detect micro-calcification in active unstable atherosclerosis.
The position of this disease as a leading cause of death worldwide is reflected in out current release, where a database search for "atherosclerosis" retrieves 28 target and ligand entries.
(1) Irkle A, Vesey AT, Lewis DY et al. (2015). Identifying active vascular microcalcification by (18)F-sodium fluoride positron emission tomography.
Nat Commun. 6: 7495. [PMID: 26151378]
A new biased apelin receptor agonist
Comments by the GtoPdb team
A new publication describes the discovery of first apelin receptor agonist biased towards the desirable positive inotropic and vasodilatory actions of the endogenous peptide but with reduced recruitment of β-arrestin, internalization and desensitization of the receptor (1). The entry for MM07 is ligand id 8523, shown below. This now joins the other ligands for the Apelin receptor.
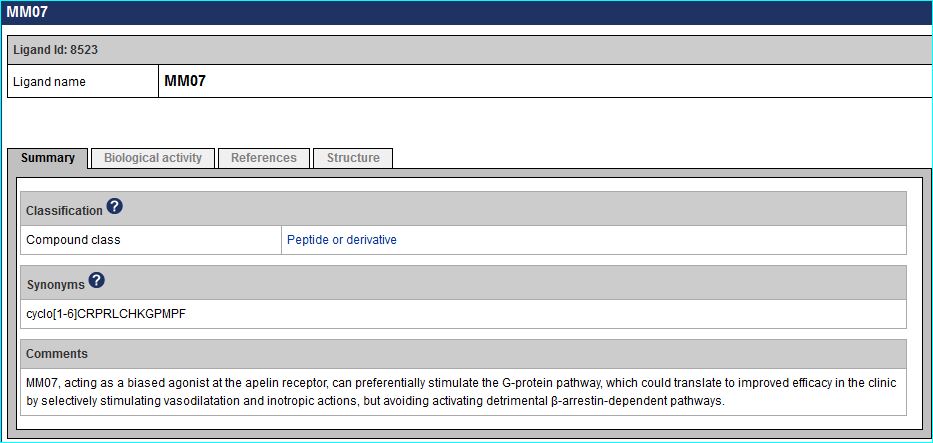
(1) Brame AL, Maguire JJ, Yang P et al. (2015). Design, characterization, and first-in-human study of the vascular actions of a novel biased apelin receptor agonist.
Hypertension. 65: 834-40. [PMID: 25712721]
Confirmed pairing of GPR139 and amino acids L-Tryptophan and L-Phenylalanine
Comments by the GtoPdb team
In 2014 the Gloriam group (that also hosts GPCRdb) published the first endogenous ligand assignment for GPR139 with L-alpha-amino acids (1). An independent confirmation of the essential amino acids L-Tryptophan and L-Phenylalanine has just appeared from Jansen R&D (2). Related Jansen data has also appeared in patent WO2014152917. In accordance with NC-IUPHAR's recommendations specified in (3), GPR139 thus meets the criteria of independent reports of endogenous ligands. If the deorphanisation is ratified at the next NC-IUPHAR meeting along with concomitant nomenclature changes proposed, we will update the comments accordingly. Meanwhile the GPR139 entry has been updated, including with recently reported surrogate small-molecule ligands, and will go live at the next release.
(1) Isberg V, Andersen KB, Bisig C et al. (2014). Computer-aided discovery of aromatic l-α-amino acids as agonists of the orphan G protein-coupled receptor GPR139.
J Chem Inf Model. 54: 1553-7. [PMID: 24826842]
(2) Liu C, Bonaventure P, Lee G et al. (2015). GPR139, an Orphan Receptor Highly Enriched in the Habenula and Septum, is Activated by the Essential Amino Acids L-Tryptophan and L-Phenylalanine.
Mol Pharmacol. 2015 Sep 8. pii: mol.115.100412. [Epub ahead of print] [PMID: 26349500]
(3) Davenport AP, Alexander SP, Sharman JL et al. (2013). International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands.
Pharmacol Rev. 65: 967-86. [PMID: 23686350]
Modelling Allosteric Modulation
Communicated via the authors from GPCRdb with comments by the GtoPdb team
A paper in Nature's Scientific Reports, "Selective Negative Allosteric Modulation Of Metabotropic Glutamate Receptors – A Structural Perspective of Ligands and Mutants" (1), reports a comprehensive comparison of the ligands in the context of their binding sites.
Relevant ligand compilations, including allosteric sections, are linked from our receptor entries for mGlu2, mGlu3 and mGlu7. The example of the mGlu3 allosteric ligand table is shown below: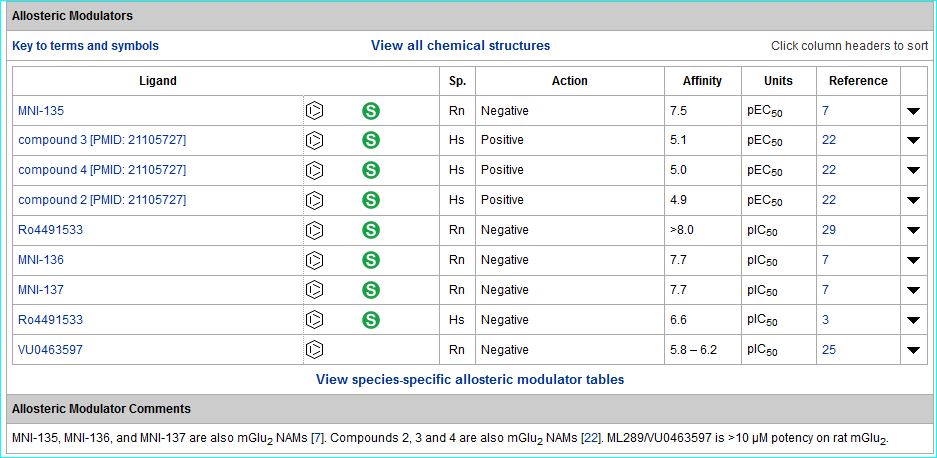
More structural information is available in the linked GPCRdb entries we include for each receptor. The sub-type selective ligands as topics in the paper are:
- FITM
- Mavoglurant
- RO5488608
- MMPIP
- We have also now added a record for ML337 with the ligand ID 8765 which will go live at our next database release.
(1) Harpsøe K, Isberg V, Tehan BG et al. (2015). Selective Negative Allosteric Modulation Of Metabotropic Glutamate Receptors – A Structural Perspective of Ligands and Mutants.
Sci Rep. 5: 13869. [PMID: 26359761]
A new approach to mapping ligandable lipid-binding proteins complemented by a knowledgebase for lipid enzymology and biology
Comments by Chris Southan
Two papers in May/June 2015 present a significant expansion in lipid metabolism and its associated prospective target landscape. The Cravatt team are pioneers in activity-based protein profiling (ABPP). In their new Cell paper (1) they adapt the approach to lipid based mass-spec labeling probes for lipid-protein binding interactions, rather than enzymatic turnover per-se. The results picked up ~ 1,000 proteins with just arachidonyl probes. These included not only many unknowns (and by implication novel lipid binding proteins of target interest) but also, unexpectedly, some known drug targets (i.e. as secondary targets). They also report a selective ligand MJN228 for a lipid-binding protein (NUCB1) that perturbs endocannabinoid and eicosanoid metabolism (the interactions entered into the database should be live in our July release). This paper was deemed worthy of comment at the popular "In the Pipeline" blog.
As an orthogonal approach to integrating lipidomic data with biological knowledge Swiss-Prot and SIB have just published their major push on lipid enzymology. Consequently, SwissLipids (2) includes over 244, 000 known and theoretical lipids, over 800 proteins, and curated links to over 620 peer-reviewed publications.
(1) Niphakis MJ, Lum KM, Cognetta AB 3rd et al. (2015). A Global Map of Lipid-Binding Proteins and Their Ligandability in Cells.
Cell. 161: 1668-80. [PMID: 26091042]
(2) Aimo L, Liechti R, Hyka-Nouspikel N et al. (2015). The SwissLipids knowledgebase for lipid biology.
Bioinformatics. 2015 May 5. pii: btv285. [Epub ahead of print] [PMID: 25943471]
Structural Basis for Receptor Activity-Modifying Protein-Dependent Selective Peptide Recognition by a G Protein-Coupled Receptor.
Comments by Anthony Davenport
Booe et al. report two crystal structures that reveal how selectivity of the GPCR, calcitonin receptor-like receptor (CLR) for two key peptides in the cardiovascular system, calcitonin gene-related peptide and adrenomedullin, is modulated by RAMP proteins.
(1) Booe JM, Walker CS, Barwell J et al. (2015). Structural Basis for Receptor Activity-Modifying Protein-Dependent Selective Peptide Recognition by a G Protein-Coupled Receptor.
Mol Cell. 58: 1040-52. [PMID: 25982113] [PDB: 4RWG, 4RWF]
Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1
Recommended by Tom Bonner
(1) Chrencik JE, Roth CB, Terakado M et al. (2015). Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1
Cell. 161: 1633-43. [PMID: 26091040]
Structure of the Angiotensin Receptor Revealed by Serial Femtosecond Crystallography
Recommended by Eliot Ohlstein
(1) Zhang H, Unal H, Gati C et al. (2015). Structure of the Angiotensin Receptor Revealed by Serial Femtosecond Crystallography
Cell. 161: 833-44. [PMID: 25913193] [PDB: 4YAY]
Calcium-sensing receptor antagonists abrogate airway hyperresponsiveness and inflammation in allergic asthma
Recommended by Rick Neubig
(1) Yarova PL, Stewart AL, Sathish V et al. (2015). Calcium-sensing receptor antagonists abrogate airway hyperresponsiveness and inflammation in allergic asthma
Sci. Transl. Med. 7: 284ra60. [PMID: 25904744]
Crystal structures of the human adiponectin receptors
Comments by Steve Alexander
Adiponectin receptors are 7-transmembrane receptors but fail to couple to G proteins. Sequence analysis suggested an inverted topology compared to classical GPCR (i.e. an intracellular C-terminus). In this paper, 2.9 and 2.4 Å resolution crystal structures of the AdipoR1 and AdipoR2 are described. Both receptors appear to have a large cavity in which a zinc atom is present, in co-ordination with histidine residues. The authors speculate that this might be associated with a lipid hydrolysis function, thereby allowing regulation of intracellular signalling pathways, such as PPARalpha.
(1) Tanabe H, Fujii Y, Okada-Iwabu M et al. (2015). Crystal structures of the human adiponectin receptors.
Nature. 520: 312-6. [PMID: 25855295] [PDB: 3WXW, 3WXV]
Crystal structures of viral chemokines and receptors provide insights into chemokine recognition
Comments by Michael Spedding
Phil Murphy established the IUPHAR chemokine receptor nomenclature in 2000 (1), and chemokines and their receptors perform important roles in host-pathogen defence, with well-established targets in the case of HIV infection, for example. There is a fast developing host versus bacteria/virus evolutionary 'arms race' which markedly affects structure of ligands and receptors, and also this means that interspecies comparisons can be difficult when there is such evolutionary pressure. This is elegantly shown in two crystal structure papers in Science where Qin et al. (2) show the human CXCR4 receptor cross-linked to the viral chemokine vMIP-II. In contrast, Burg et al. (3) show how the human chemokine CX3CL1 interacts with the virally-encoded US28 receptor. The crystal structures can help the design of new antiviral agents, and show that not all receptors are 'endogenous'.
For more information also read the perspective in Science (4).
(1) Murphy PM, Baggiolini M, Charo IFet al. (2000). International Union of Pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol. Rev. 52: 145-176. [Full text]
(2) Qin L, Kufareva I, Holden LG et al. (2015). Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine.
Science. 347: 1117-22. [Full text] [PDB: 4RWS]
(3) Burg JS, Ingram JR, Venkatakrishnan AJ et al. (2015). Structural basis for chemokine recognition and activation of a viral G protein–coupled receptor.
Science. 347: 1113-17. [Full text]
(4) Standfuss J (2015). Viral chemokine mimicry.
Science. 347: 1071-72. [Full text]
SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1
Comments by Stephen Alexander
The SoLute Carrier (SLC) family of transporters are associated with movement of solutes across membranes driven by ion gradients. The SLC38 family includes 11 transporters, where two groups of cell-surface transporters are defined, equivalent to system A and system N sodium-dependent amino acid transporters. A further group of six transporters in the SLC38 family have no ascribed function, and so are designated as orphans.
This report describes SLC38A9, one of those orphans, which the authors suggest is a lysosomal transporter able to transport 3H-glutamine, and to a lesser extent, 3H-arginine and 3H-asparagine. Of particular interest is the apparent ability of this transporter to enhance the activity of mTORC1, a nexus for the regulation of cellular metabolism, including protein synthesis. The authors suggest that this transporter may, therefore, represent a mechanism for integration of protein turnover, and hence cell growth and proliferation.
(1) Rebsamen M, Pochini L, Stasyk T et al. (2015). SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1.
Nature. 2015 Jan 7. doi: 10.1038/nature14107. [Epub ahead of print] [PMID: 25561175]
Generic GPCR residue numbers – aligning topology maps while minding the gaps
A new version of the generic GPCR residue numbering system is discussed along with information on using GPCRDB web tools to number any receptor sequence or structure.
(1) Isberg V, de Graaf C, Bortolato A, Cherezov V, Katritch V, Marshall FH, Mordalski S, Pin J, Stevens RC, Vriend G, Gloriam DE (2015). Generic GPCR residue numbers - aligning topology maps while minding the gaps.
Trends Pharmacol Sci. 36: 22-31. [PMID: 25541108]
Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant
Recommended by Tom Bonner
(1) Yin J, Mobarec JC, Kolb P, Rosenbaum DM. (2014). Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant
Nature. Dec 22. doi: 10.1038/nature14035 [Epub ahead of print] [PMID: 25533960] [PDB: 4RNB]
2014
X-ray structure of the mGlu5 receptor
Comments by Fiona Marshall
The team from Heptares have published the structure of the transmembrane domain of the mGlu5 receptor in Nature (1). The structure is in complex with the negative allosteric modulator (NAM) mavoglurant which is currently in clinical trials for OCD and depression and has previously been evaluated in Fragile X disorder. The overall structure is similar to the recently published structure of the related mGlu1 receptor (2) however the ligand binds much deeper within the transmembrane domain. The interactions of mavoglurant with the allosteric binding pocket provide evidence of its mechanism of action as a NAM via the stabilisation of a water network which links transmembrane helices (TM) 3, 6 and 7. The structure also explains why small chemical changes to allosteric modulators can result in 'mode switching' between NAMs and PAMs. This new structure will enable structure based design methods to be applied to this receptor and others in the Class C sub family of GPCRs.
The paper also includes a detailed comparison of the structural features of Class C receptors with those of Class A and Class B. Motifs which are conserved within Class C are compared to those with similar functions in the other classes – for example the ionic lock which anchors TM3 and TM6 regulating receptor activation. This is complemented with mutagenesis experiments which further explore the nature of these structural features and their role in receptor activation and signalling.
(1) Doré, A, Okrasa K, Patel J, Serrano-Vega M, Bennett K, Cooke R, Errey J, Jazayeri A, Khan S, Tehan B, Weir M, Wiggin G and Marshall FH. (2014). Structure of a class C GPCR metabotropic glutamate receptor 5 transmembrane domain.
Nature. 2014 Jul 6. doi:10.1038/nature13396. [Full text] [PDB: 4OO9]
(2) Wu H, Wang C, Gregory KJ, Han GW, Cho HP, Xia Y, Niswender CM, Katritch V, Meiler J, Cherezov V, Conn PJ, Stevens RC. (2014). Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator.
Science. 344: 58-64. [PMID: 24603153]
Crystal structure of a human GABAA receptor
Recommended by Chris Southan
(1) Miller PS, Aricescu AR. (2014). Crystal structure of a human GABAA receptor.
Nature. 2014 Jun 8 doi: 10.1038/nature13293. [Epub ahead of print] [PMID: 24909990] [PDB: 4COF]
Crystal structure of GluN1/GluN2B NMDA receptor
Comments by David Wyllie
The crystal structure of a diheteromeric NMDA receptor composed of GluN1 and GluN2B subunits has been solved (1). Overall the tetrameric structure is similar to that reported for a homomeric GluA2 AMPA receptor (2) in that it is assembled as a dimer of dimers with subunits in a 1-2-1-2 orientation with a pseudo-symmetry mismatch between transmembrane and extracellular domains and a swapping of the pair of dimers between the amino terminal and ligand binding domains. In this respect, in the NMDA receptor complex GluN1 subunits can be considered to be similar to the A-C subunit and GluN2B subunits similar to the B-D subunits of the GluA2 AMPA receptor complex. Nevertheless there are intriguing differences between the two receptor structures. For example, the amino and ligand binding domains in the NMDA receptor structure are considerably more compact than that observed in the GluA2 AMPA receptor and there are sites of interaction both within subunit and between subunits which are present in the NMDA receptor structure but which are absent in the AMPA receptor. These sites of interaction are suggested to relate to the fact that NMDA receptors are regulated by a very much greater number of allosteric modulators than their AMPA receptor counterparts and are likely to inform future studies aimed at developing NMDA receptor subtype-selective ligands.
(1) Karakas E, Furukawa H. (2014). Crystal structure of a heterotetrameric NMDA receptor ion channel.
Science. 344: 992-7. [PMID: 24876489] [PDB: 4PE5]
(2) Sobolevsky AI, Rosconi MP, Gouaux E. (2009). X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor.
Nature. 462: 745-56. [PMID: 19946266] [PDB: 3KGC, 3KG2]
Update: A second GluN1-GluN2B NMDA receptor structure from Xenopus laevis has been published in Nature (3) with commentary (4).
(3) Lee C-H, Lü W, Carlisle Michel J, Goehring A, Du J, Song X, Gouaux E. (2014). NMDA receptor structures reveal subunit arrangement and pore architecture.
Nature. 511: 191-7. [Full text]
(4) Stroebel D, Paoletti P (2014). Neuroscience: A structure to remember.
Nature. 511: 162-3. [Full text]
The Sphingolipid Receptor S1PR2 Is a Receptor for Nogo-A Repressing Synaptic Plasticity.
Recommended by Rick Neubig
Click here for database entry: S1P2 receptor
(1) Kempf A, Tews B, Arzt ME, Weinmann O, Obermair FJ, Pernet V, Zagrebelsky M, Delekate A, Iobbi C, Zemmar A, Ristic Z, Gullo M, Spies P, Dodd D, Gygax D, Korte M, Schwab ME. (2014) The Sphingolipid Receptor S1PR2 Is a Receptor for Nogo-A Repressing Synaptic Plasticity.
PLoS Biol. 12: e1001763 [PMID: 24453941]
Serial Femtosecond Crystallography of G Protein-Coupled Receptors
Comments by Anthony Harmar
An x-ray free-electron laser (XFEL) producing individual 50-femtosecond-duration x-ray pulses was used to obtain a high-resolution structure of the human 5-HT2B receptor, bound to the agonist ergotamine, at room temperature. Compared with the structure solved by using traditional microcrystallography (2), the room-temperature XFEL structure probably includes features that more accurately represent the receptor structure and dynamics in a cellular environment. For example, a salt bridge between helices V and VI was identified.
(1) Liu W, Wacker D, Gati C, Han GW, James D, Wang D, Nelson G, Weierstall U, Katritch V, Barty A, Zatsepin NA, Li D, Messerschmidt M, Boutet S, Williams GJ, Koglin JE, Seibert MM, Wang C, Shah ST, Basu S, Fromme R, Kupitz C, Rendek KN, Grotjohann I, Fromme P, Kirian RA, Beyerlein KR, White TA, Chapman HN, Caffrey M, Spence JC, Stevens RC, Cherezov V. (2013). Serial femtosecond crystallography of G protein-coupled receptors.
Science. 342: 1521-4. [PMID: 24357322] [PDB: 4NC3]
(2) Wacker D, Wang C, Katritch V, Han GW, Huang XP, Vardy E, McCorvy JD, Jiang Y, Chu M, Siu FY, Liu W, Xu HE, Cherezov V, Roth BL, Stevens RC. (2013). Structural features for functional selectivity at serotonin receptors.
Science. 340: 615-9. [PMID: 23519215] [PDB: 4IB4]
2013
Activation and allosteric modulation of a muscarinic acetylcholine receptor.
Recommended by Tom Bonner
Click here for database entry: M2 receptor
(1) Kruse AC, Ring AM, Manglik A, Hu J, Hu K, Eitel K, Hübner H, Pardon E, Valant C, Sexton PM, Christopoulos A, Felder CC, Gmeiner P, Steyaert J, Weis WI, Garcia KC, Wess J, Kobilka BK. (2013) Activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nature. 2013 Nov 20. doi: 10.1038/nature12735. [Epub ahead of print] [PMID: 24256733]
Structure of the human CCR5 chemokine receptor bound to the marketed HIV drug maraviroc.
Recommended by Tom Bonner
Click here for database entry: CCR5
(1) Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, Li T, Ma L, Fenalti G, Li J, Zhang W, Xie X, Yang H, Jiang H, Cherezov V, Liu H, Stevens RC, Zhao Q, Wu B. (2013) Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex.
Science. 341: 1387-90 [PMID: 24030490]
The role of Melanocortin-2 receptor accessory proteins
Comments by Anthony Harmar:
Melanocortin-2 receptor accessory proteins (MRAP AND MRAP2) are small single–transmembrane domain proteins that colocalise with melanocortin receptors in the endoplasmic reticulum and plasma membrane and are thought to play a role in the processing, trafficking and/or function of these receptors (1). Mutations in MRAP have been shown to be the cause of familial glucocorticoid deficiency type 2, a rare autosomal recessive disorder characterised by high plasma ACTH levels but severe cortisol deficiency (1,2). Normally, plasma glucocorticoid concentrations are regulated by ACTH, which stimulates MC2 receptors in the adrenal cortex to promote corticosteroid synthesis. In the absence of MRAP, MC2 receptors cannot translocate from the endoplasmic reticulum to the plasma membrane and ACTH-induced signalling is extinguished. To explore the function of the related protein MRAP2, Asai et al. (2013) characterised mice with a targeted deletion of Mrap2 and observed that they developed severe obesity at a young age (3). They showed that MRAP2 interacts with the MC4 receptor, a protein previously implicated in mammalian obesity, to enhance the responsiveness of the receptor to α-MSH. Mice with tissue-specific knockout of Mrap2 in the paraventricular hypothalamus (PVH) and a subpopulation of amygdala neurons - implicated in mediating the actions of the MC4 receptor on food intake but not on energy expenditure (4)- became similarly obese to global knockout animals. Sequencing MRAP2 in a cohort of humans with severe, early-onset obesity identified four rare, potentially pathogenic genetic variants in MRAP2, suggesting that the gene may contribute to body weight regulation in humans.
This work has several implications for the development of drugs with actions at melanocortin receptors. Firstly, expression of these receptors in transfected cell lines has been problematic, making it difficult to develop robust assays for new ligands. Co-expression of receptors with MRAPs may make this easier. Secondly, although mutations in MRAPs are a very rare cause of obesity, understanding their mechanisms of action may indicate new approaches to drug development. Thirdly, drugs with an allosteric action on complexes between receptors, MRAPs and other accessory proteins (5) may enable cell type specific and or activity-specific activation of receptors, with potential novel therapeutic benefit. Finally, the new work adds another member to a growing list of proteins that can interact with GPCRs to modulate their cellular distribution, pharmacology and/or signaling (6).
(1) Metherell LA, Chapple JP, Cooray S, David A, Becker C, Ruschendorf F et al. (2005). Mutations in MRAP, encoding a new interacting partner of the ACTH receptor, cause familial glucocorticoid deficiency type 2.
Nature genetics. 37: 166-170 [PMID: 15654338]
(2) Modan-Moses D, Ben-Zeev B, Hoffmann C, Falik-Zaccai TC, Bental YA, Pinhas-Hamiel O et al. (2006) Unusual presentation of familial glucocorticoid deficiency with a novel MRAP mutation.
J Clin Endocrinol Metab. 91: 3713-3717 [PMID: 16868047]
(3) Asai M, Ramachandrappa S, Joachim M, Shen Y, Zhang R, Nuthalapati N et al. (2013) Loss of function of the melanocortin 2 receptor accessory protein 2 is associated with mammalian obesity.
Science. 341: 275-278 [PMID: 23869016]
(4) Balthasar N, Dalgaard LT, Lee CE, Yu J, Funahashi H, Williams T et al. (2005) Divergence of melanocortin pathways in the control of food intake and energy expenditure.
Cell. 123: 493-505 [PMID: 16269339]
(5) Breit A, Buch TR, Boekhoff I, Solinski HJ, Damm E, Gudermann T. (2011) Alternative G protein coupling and biased agonism: new insights into melanocortin-4 receptor signalling.
Mol Cell Endocrinol. 331: 232-240 [PMID: 20674667]
(6) Ritter SL, Hall RA. (2009) Fine-tuning of GPCR activity by receptor-interacting proteins.
Nat Rev Mol Cell Biol. 10: 819-30 [PMID: 19935667]
X-ray structures of family B GPCRs.
In 2012 Heptares reported that they had solved the X-ray crystal structure of the first family B GPCR. The structure of the corticotropin-releasing factor receptor 1 has now been published online ahead of print in Nature (1), along with a second paper describing the structure of the human glucagon receptor (2).
(1) Hollenstein K, Kean J, Bortolato A et al. (2013) Structure of class B GPCR corticotropin-releasing factor receptor 1.
Nature. 2013 Jul 17. doi:10.1038/nature12357. [Epub ahead of print] [FULL TEXT]
(2) Sui FY, de Graaf C, Han GW et al. (2013) Structure of the human glucagon class B G-protein-coupled receptor.
Nature. 2013 Jul 17. doi:10.1038/nature12393. [Epub ahead of print] [FULL TEXT]
X-ray structure of the mammalian GIRK2–βγ G-protein complex.
Recommended by Rick Neubig
Click here for database entry: Kir3.2
(1) Whorton MR, Mackinnon R. (2013) X-ray structure of the mammalian GIRK2–βγ G-protein complex.
Nature. 2013 Jun 5. doi: 10.1038/nature12241. [Epub ahead of print] [PMID: 23739333]
The cells and circuitry for itch responses in mice.
Recommended by Anthony Harmar
(1) Mishra SK, Hoon MA. (2013) The cells and circuitry for itch responses in mice
Science. 340: 968-71 [PMID: 23704570]
Betatrophin: A Hormone that Controls Pancreatic β Cell Proliferation.
Recommended by Anthony Harmar
The authors identify a hormone, betatrophin, that is primarily expressed in liver and fat. and promotes pancreatic β cell proliferation, expands β cell mass and improves glucose tolerance (1). The hormone, the product of the C19orf80 gene, has previously been named "lipasin" because of its effects in increasing serum triglyceride levee and inhibiting lipoprotein lipase activity (2). Betatrophin is a member of the family of angiopoietin-like proteins (ANGPTLs), a family of seven secreted glycoproteins (3).
(1) Yi P, Park JS, Melton DA. (2013) Betatrophin: A Hormone that Controls Pancreatic β Cell Proliferation.
Cell. 153: 747-58 [PMID: 23623304]
(2) Zhang R. (2012) Lipasin, a novel nutritionally-regulated liver-enriched factor that regulates serum triglyceride levels.
Biochem Biophys Res Commun. 424: 786-92 [PMID: 22809513]
(3) Quagliarini F, Wang Y, Kozlitina J, Grishin NV, Hyde R, Boerwinkle E, Valenzuela DM, Murphy AJ, Cohen JC, Hobbs HH. (2012) Atypical angiopoietin-like protein that regulates ANGPTL3.
Proc Natl Acad Sci U S A. 109: 19751-6 [PMID: 23150577]
Genetic variants in GPR126 are associated with adolescent idiopathic scoliosis.
Recommended by Tom Bonner
A paper published online ahead of print in Nature Genetics shows that genetic variants in GPR126 are associated with adolescent idiopathic scoliosis (1). Supplementary figure 3 shows GPR126 expression in 20 human tissues with cartilage showing the highest abundance followed by placenta, liver and bone. The paper also quotes papers reporting GPR126-null mice having limb posture abnormalities and growth failure as well as deficits in peripheral nerve development and myelination (2) and being essential for myelination in zebrafish (3).
(1) Kou I, Takahashi Y, Johnson TA, Takahashi A, Guo L, Dai J, Qiu X, Sharma S, Takimoto A, Ogura Y, Jiang H, Yan H, Kono K, Kawakami N, Uno K, Ito M, Minami S, Yanagida H, Taneichi H, Hosono N, Tsuji T, Suzuki T, Sudo H, Kotani T, Yonezawa I, Londono D, Gordon D, Herring JA, Watanabe K, Chiba K, Kamatani N, Jiang Q, Hiraki Y, Kubo M, Toyama Y, Tsunoda T, Wise CA, Qiu Y, Shukunami C, Matsumoto M, Ikegawa S. (2013) Genetic variants in GPR126 are associated with adolescent idiopathic scoliosis.
Nat Genet. 45: 676-679 [PMID: 23666238]
(2) Monk KR, Oshima K, Jörs S, Heller S, Talbot WS. (2011) Gpr126 is essential for peripheral nerve development and myelination in mammals.
Development. 138: 2673–2680 [PMID: 21613327]
(3) Monk KR, Naylor SG, Glenn TD, Mercurio S, Perlin JR, Dominguez C, Moens CB, Talbot WS. (2009) A G protein–coupled receptor is essential for Schwann cells to initiate myelination.
Science. 325: 1402–1405 [PMID: 19745155]
Update to the G Protein-Coupled Receptor list: recommendations for new pairings with cognate ligands
IUPHAR review article published in the journal Pharmacological Reviews updating the G Protein-Coupled Receptor list with recommendations for new pairings with cognate ligands. Click here to access the database pages for orphan GPCRs.
Davenport AP, Alexander SP, Sharman JL, Pawson AJ, Benson HE, Monaghan AE, Liew WC, Mpamhanga CP, Bonner TI, Neubig RR, Pin JP, Spedding M, Harmar AJ. (2013)
International Union of Basic and Clinical Pharmacology. LXXXVIII. G Protein-Coupled Receptor List: Recommendations for New Pairings with Cognate Ligands.
Pharmacol Rev. 65: 967-86. [Abstract] [Full text]
Evidence for testicular orphan nuclear receptor 4 in the etiology of Cushing disease
Recommended by Rick Neubig
A paper published online ahead of print in PNAS describes evidence for the testicular orphan nuclear receptor 4 (TR4, nuclear receptor subfamily 2, group C, member 2) in the etiology of Cushing disease.
(1) Du L, Bergsneider M, Mirsadraei L, Young SH, Jonker JW, Downes M, Yong WH, Evans RM, Heaney AP. (2013) Evidence for orphan nuclear receptor TR4 in the etiology of Cushing disease.
PNAS. 2013 May 7 doi: 10.1073/pnas.1306182110 [Epub ahead of print] [Abstract]
Structure of the human smoothened receptor bound to an antitumour agent
Recommended by Anthony Harmar
A paper published online ahead of print in Nature describes the crystal structure of the transmembrane domain of the human SMO receptor bound to the small-molecule antagonist LY2940680 at 2.5 Å resolution.
(1) Wang C, Wu H, Katritch V, Han GW, Huang XP, Liu W, Siu FY, Roth BL, Cherezov V, Stevens RC. (2013) Structure of the human smoothened receptor bound to an antitumour agent.
Nature. 2013 May 1. doi: 10.1038/nature12167. [Epub ahead of print] [PMID: 23636324]
Crystal structures of arrestin
Recommended by Tom Bonner
Two papers published online ahead of print in Nature describe the crystal structures of arrestin. Read the commentary in Nature here.
(1) Kim YJ, Hofmann KP, Ernst OP, Scheerer P, Choe HW, Sommer ME. (2013) Crystal structure of pre-activated arrestin p44.
Nature. 497: 142-6 [PMID: 23604253]
(2) Shukla AK, Manglik A, Kruse AC, Xiao K, Reis RI, Tseng WC, Staus DP, Hilger D, Uysal S, Huang LY, Paduch M, Tripathi-Shukla P, Koide A, Koide S, Weis WI, Kossiakoff AA, Kobilka BK, Lefkowitz RJ. (2013) Structure of active β-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide.
Nature. 497: 137-41 [PMID: 23604254]
A pharmacological organization of G protein-coupled receptors
Recommended by Anthony Davenport
(1) Lin H, Sassano MF, Roth BL, Shoichet BK. (2013) A pharmacological organization of G protein-coupled receptors.
Nat Methods. 10: 140-6. [PMID: 23291723]
Crystal structures of the 5-HT1B and 5-HT2B G protein-coupled receptors
Recommended by Tom Bonner
Two papers published online ahead of print in Science describe the crystal structures of the serotonin G protein-coupled receptors 5-HT1B and 5-HT2B.
(1) Wang C, Jiang Y, Ma J, Wu H, Wacker D, Katritch V, Han GW, Liu W, Huang XP, Vardy E, McCorvy JD, Gao X, Zhou EX, Melcher K, Zhang C, Bai F, Yang H, Yang L, Jiang H, Roth BL, Cherezov V, Stevens RC, Xu HE. (2013) Structural Basis for Molecular Recognition at Serotonin Receptors.
Science. 2013 Mar 21 [Epub ahead of print] [PMID: 23519210]
(2) Wacker D, Wang C, Katritch V, Han GW, Huang XP, Vardy E, McCorvy JD, Jiang Y, Chu M, Siu FY, Liu W, Xu HE, Cherezov V, Roth BL, Stevens RC. (2013) Structural Features for Functional Selectivity at Serotonin Receptors.
Science. 2013 Mar 21 [Epub ahead of print] [PMID: 23519215]
A screen of 10000 ligands against 82 GPCRs confirms pairings of cognate ligands with orphan receptors and identifies novel surrogate ligands
An article published online ahead of print in the Journal of Biomolecular Screening reports the results of screening ~10,000 ligands against eighty-two G protein-coupled receptors, mainly orphans, using the PathHunter β-arrestin recruitment assays. Pairings of cognate ligands with orphan receptors were confirmed and a number of novel surrogate ligands identified.
Southern C, Cook JM, Neetoo-Isseljee Z, Taylor DL, Kettleborough CA, Merritt A, Bassoni DL, Raab WJ, Quinn E, Wehrman TS, Davenport AP, Brown AJ, Green A, Wigglesworth MJ, Rees S. (2013)
Screening β-Arrestin Recruitment for the Identification of Natural Ligands for Orphan G-Protein-Coupled Receptors.
J Biomol Screen. doi: 10.1177/1087057113475480 [Epub ahead of print] [PMID: 23396314]
IUPHAR review article published in the journal Pharmacological Reviews on the nomenclature and pharmacology of the Complement Peptide receptors.
Klos A, Wende E, Wareham KJ, Monk PN. (2013)
International Union of Pharmacology. LXXXVII. Complement Peptide C5a, C4a, and C3a Receptors.
Pharmacol Rev. 65: 500-543. [Abstract] [Full text]
The second IUPHAR review article to be published in the British Journal of Pharmacology is "New concepts in pharmacological efficacy at 7TM receptors" by Terry Kenakin.
Kenakin T. (2013)
New concepts in pharmacological efficacy at 7TM receptors: IUPHAR Review 2.
Pharmacol Rev. 65: 500-543. [Abstract] [Full text]
Anti-inflammatory lipoxin A4 is an endogenous allosteric enhancer of CB1 cannabinoid receptor.
Recommended by Rick Neubig
A recent article published in the journal PNAS shows that the anti-inflammatory lipid lipoxin A4 is an endogenous allosteric enhancer of the CB1 cannabinoid receptor.
Pamplona FA, Ferreira J, Menezes de Lima O Jr, Duarte FS, Bento AF, Forner S, Villarinho JG, Bellochio L, Wotjak CT, Lerner R, Monory K, Lutz B, Canetti C, Matias I, Calixto JB, Marsicano G, Guimarães MZ, Takahashi RN. (2012)
Anti-inflammatory lipoxin A4 is an endogenous allosteric enhancer of CB1 cannabinoid receptor.
Proc Natl Acad Sci U S A. 109: 21134-9. [PMID: 23150578]
2012
Orphan receptor GPR107 identified as the target of the novel neuropeptide neuronostatin.
Comments by Rick Neubig:
This paper uses an interesting approach of profiling a series of cell lines with or without responsiveness to neuronostatin for expression of orphan receptors. All lines that were responsive expressed a set of 4 orphan receptors. si-RNA-mediated Knock down only of GPR107 prevented neuronstatin-stimulated c-fos mRNA expression. Similar loss-of-function studies were done with antisense oligos in vivo. These results suggest that GPR107 is the target of the novel neuropeptide neuronostatin.
(1) Yosten GL, Redlinger LJ, Samson WK. (2012)
Evidence for an interaction of neuronostatin with the orphan G protein-coupled receptor, GPR107.
Am J Physiol Regul Integr Comp Physiol. 303 (9): R941-9. [PMID: 22933024]
2012 Nobel prize for Chemistry awarded for work on G protein-coupled receptors.
Recommended by Anthony Davenport
The 2012 Nobel prize for Chemistry has been awarded to Robert Lefkowitz and Brian Kobilka for their work on G protein-coupled receptors (GPCRs). Congratulations to Professors Lefkowitz and Kobilka! Click here for the scientific background on this award.
Heptares solves first family B GPCR structure.
Recommended by Anthony Davenport
Heptares solves X-ray crystal structure of the first family B GPCR.
Structure of the chemokine receptor CXCR1.
Recommended by Tom Bonner
A paper in Nature describes the structure of the chemokine receptor CXCR1 in phospholipid bilayers. This is important as it is the first structure of an unmodified GPCR and first structure in a lipid bilayer. It is also the first structure to be determined by NMR spectroscopy.
(1) Park SH, Das BB, Casagrande F, Tian Y, Nothnagel HJ, Chu M, Kiefer H, Maier K, De Angelis AA, Marassi FM, Opella SJ. (2012)
Structure of the chemokine receptor CXCR1 in phospholipid bilayers.
Nature. 491: 779-83. [PMID: 23086146]
Update article on IUPHAR-DB published in Nucleic Acids Research.
An update paper describing new content and features of the IUPHAR Database is to be published in the 2013 Nucleic Acids Research Database Issue. The paper is available online with advance access.
Sharman JL, Benson HE, Pawson AJ, Lukito V, Mpamhanga CP, Bombail V, Davenport AP, Peters JA, Spedding M, Harmar AJ, and NC-IUPHAR. (2013)
IUPHAR-DB: updated database content and new features.
Nucl. Acids Res. (Database Issue). 2012 Oct 18. [Epub ahead of print] [Abstract] [Full text]
IUPHAR review article published on the nomenclature, function and pharmacology of Orexin receptors.
Gotter AL, Webber AL, Coleman PJ, Renger JJ, Winrow CJ. (2012)
International Union of Basic and Clinical Pharmacology. LXXXVI. Orexin Receptor Function, Nomenclature and Pharmacology.
Pharmacol Rev. 64: 389-420. [Abstract] [Full text]
How to use the IUPHAR Database: step-by-step protocol and examples
The IUPHAR Database team has produced a comprehensive protocol describing how to use the IUPHAR Database for Receptor Binding Techniques published by Springer Protocols, with step-by-step instructions, screenshots, examples and tips to help users make the most of the database.
Mpamhanga CP, Sharman JL, Harmar AJ, and NC-IUPHAR. (2012)
How to Use the IUPHAR Receptor Database to Navigate Pharmacological Data.
Methods Mol Biol. 897: 15-29. In Receptor Binding Techniques edited by Anthony P. Davenport (Springer Protocols). [PMID: 22674159]
IUPHAR review article published on the pharmacology and functions of VIP and PACAP receptors.
The first in a new series of IUPHAR Review articles in British Journal of Pharmacology.
Harmar AJ, Fahrenkrug J, Gozes I, Laburthe M, May V, Pisegna JR, Vaudry D, Vaudry H, Waschek JA, Said SI. (2012)
IUPHAR Reviews 1: Pharmacology and functions of receptors for vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide.
Br J Pharmacol. 166: 4-17. [PMID: 22289055]
Crystal structures of the μ and κ opioid receptors
Comments by Brian M. Cox:
It is now almost 60 years since Beckett and Casy first proposed that morphine and related drugs must act through a specific receptor in brain to induce analgesia (1), and nearly 40 years since three groups independently showed the presence of high affinity binding sites for such drugs in the central nervous system (2, 3, 4). Another milestone in our understanding of the actions of morphine like drugs comes with the publication this month of the crystal structures of two of the four closely related opioid peptide receptors, the κ opioid receptor (5) and the μ opioid receptor (6). Morphine and other opiates used therapeutically act predominantly through the μ receptor while the κ receptor is activated predominantly by some ketocyclazocines, by the hallucinogenic agent salvinorin A, and by the endogenous opioid dynorphin.
The new reports follow closely on reports earlier this year of other GPCRs. The two groups responsible for these latest developments used similar strategies; the receptors were crystallized as complexes with very tightly binding highly receptor-type-selective antagonist ligands; JDTic in the case of the κ receptor and β-FNA for the μ receptor. Thus in each case the receptor is visualized in an inactive conformation. Nevertheless, some interesting features are immediately apparent. Both receptors crystallized as dimers, with more than one potential interface between adjacent receptor monomers as possible dimerization sites. Higher polymerization states and heterodimerization with other GPCRs are possible. These observations provide a structural basis for earlier proposals that opioid receptors might function as dimers or higher polymers (7). Opiate drugs are also known for their rapid reversibility - the immediacy of the reversal of opiate-induced respiratory depression by naloxone can be dramatic. The new studies show that the ligand binding pockets of both the μ and κ receptors are unusually exposed or open relative to other GPCRs. The accessibility of the binding pocket favors rapid dissociation (except in the case of irreversible antagonists such as β-FNA). Since the affinity of many agonist and antagonists at μ or κ receptors is high despite their rapid reversibility, their association rates must also be very high.
Another feature of opioid receptors is the apparent ability of different ligands acting through the same receptor type to direct signaling through different effector pathways. Ligands for opioid receptors are chemically very heterogeneous. The reported structures for the μ and κ receptors point to accessory sites around the common ligand binding pocket for each receptor that provide additional points of receptor interaction for some ligands. Much work needs to be done to understand the basis of agonism at these receptors, but it is tempting to speculate that these additional interaction sites for some ligands might be exploited in the design of agonists preferentially driving signaling through alternative transduction pathways.
(1) Beckett AH and Casy AF. (1954) Synthetic analgesics: stereochemical considerations.
J Pharm Pharmacol. 12: 986-1001. [PMID: 13212680]
(2) Pert CB and Snyder SH. (1973) Opiate receptor: demonstration in nervous tissue.
Science. 179: 1011-1014. [PMID: 4687585]
(3) Simon EJ, Hiller JM, Edelman I. (1973) Stereospecific binding of the potent narcotic analgesic (3H)etorphine to rat brain homogenate.
Proc Natl Acad Sci USA. 70: 1947-1949. [PMID: 4516196]
(4) Terenius L. (1973) Stereospecific interaction between narcotic analgesics and a synaptic plasma membrane fraction of rat cerebral cortex.
Acta Pharmacol Toxicol. 32: 317-320. [PMID: 4801733]
(5) Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, Mascarella SW, Westkaemper RB, Mosier PD, Roth BL, Cherezov V, Stevens RC. (2012) Structure of the human κ-opioid receptor in complex with JDTic.
Nature. doi: 10.1038/nature10939. [Epub ahead of print] [PMID: 22437504]
(6) Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. (2012) Crystal structure of the μ-opioid receptor bound to a morphinan antagonist.
Nature. doi: 10.1038/nature10954. [Epub ahead of print] [PMID: 22437502]
(7) Jordan BA and Devi L. (1999) G-protein-coupled receptor heterodimerization modulates receptor function.
Nature. 399: 697-700. [PMID: 10385123]
Crystal structure of the S1P1 receptor
Comments by Tony Harmar and Jerold Chun:
The structure of the S1P1 receptor fused with T4 lysozyme, in complex with a selective antagonist sphingolipid mimic (ML056), has been reported at 2.8 Å and 3.35 Å resolution by scientists from the Scripps Research Institute and their drug discovery company Receptos (San Diego, CA). This approach that has revealed important structures of other inactive conformations of GPCRs unbound to G proteins. The structure of the ligand-binding pocket of the receptor suggests that there is limited access for ligand from the extracellular surface of the receptor; rather, ligands may gain access to the binding pocket from within the membrane bilayer, as has been proposed for retinal loading into opsin and for the entry of anandamide into cannabinoid receptors. Modeling and site-directed mutagenesis studies led to the mapping of a putative binding pocket for a subclass of agonists that is distinct from the putative binding site for the endogenous ligand. The active metabolite of the S1P1 agonist fingolimod, which actually appears to involve efficacy through functional antagonistic properties on lymphocytes and CNS cells, represents the first oral therapy for multiple sclerosis, and several other compounds are in development that possess S1P1 modulatory activities. Structural data on the critical signaling complex of S1P1 with its biologically relevant heterotrimeric G proteins, as has been reported for the structure of the β2 adrenergic receptor in complex with Gs, await further studies.
(1) Hanson MA, Roth CB, Jo E, Griffith MT, Scott FL, Reinhart G, Desale H, Clemons B, Cahalan SM, Schuerer SC, Sanna MG, Han GW, Kuhn P, Rosen H, Stevens RC. (2012)
Crystal Structure of a Lipid G Protein–Coupled Receptor
Science. 335: 851-5. [PMID: 22344443]
Structures of M2 and M3 muscarinic acetylcholine receptors
Comments by A.J. Harmar:
The structures of the human M2 receptor and the rat M3 receptor, each in a complex with an inverse agonist (3-quinuclidinyl-benzilate and tiotropium, respectively) have been reported (1,2). In each case, the third intracellular loop of the receptor was replaced with T4 lysozyme – an approach that has been used to obtain crystal structures of several other GPCRs. The overall structures of the two receptors are similar, even in some regions (e.g. intracellular and extracellular loops) that display divergent amino acid sequences. In both cases, there is a “large extracellular vestibule as part of an extended hydrophilic channel containing the orthosteric ligand binding site” – a feature that has not been seen in previous GPCR structures. The orthosteric ligand binding sites share many common features with other unrelated acetylcholine binding proteins. Amino acid residues forming the binding pocket are highly conserved between muscarinic receptor subtypes, explaining why receptor subtype specific orthosteric ligands have been difficult to obtain. However, the new structures demonstrate some differences between the binding sites in M2 and M3 receptors that might permit the development of subtype-selective ligands.
There are significant differences in the position of the cytoplasmic end of TM5 and of ICL2 between the two receptors, which may contribute to their different G protein coupling specificities: the position of TM5 was similar in the Gi/o coupled M2, D3 and CXCR4 receptors, whereas the Gq/11-coupled M3 and H1 receptors exhibit a different conformation. A better understanding of G protein coupling specificity will require solution of the structures of more receptor – G protein complexes, as has been achieved for the β2 adrenoceptor – Gs complex (3).
Simulations of the binding of tiotropium to M2 and M3 receptors showed that the ligand pauses at a known allosteric site during association and dissociation from the receptor, leading the authors to suggest that “conceivably, therapeutic molecules could be rationally engineered to act independently as both allosteric and orthosteric ligands”.
(1) Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T. (2012)
Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist.
Nature. 482: 547-51. [PMID: 22278061]
(2) Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, Shaw DE, Weis WI, Wess J, Kobilka BK. (2012)
Structure and dynamics of the M3 muscarinic acetylcholine receptor.
Nature. 482: 552-6. [PMID: 22358844]
(3) Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. (2011)
Crystal structure of the β2 adrenergic receptor-Gs protein complex.
Nature. 477: 549-55. [PMID: 21772288]
2011
IUPHAR review article published on the Calcium-Activated Chloride Channels.
Huang F, Wong X, Jan LY. (2012) International Union of Basic and Clinical Pharmacology. LXXXV: Calcium-Activated Chloride Channels.
Pharmacol Rev. 64: 1-15. [PMID: 22090471]
N.B. This family is not currently listed in IUPHAR-DB. See the Guide to Receptors and Channels (GRAC) page on Calcium-Activated Chloride Channels.
The 7-transmembrane receptor LGR5: A GPCR no more?
Comments by Elizabeth R. Lawlor and Richard R. Neubig, University of Michigan, Ann Arbor, Michigan
LGR5 and its close relatives LGR4 and LGR6 were first identified as a family of structurally distinct 7-transmembrane receptors with homology to glycoprotein hormone receptors. Characterized by large N-terminal extracellular domains comprised of 17 leucine-rich repeats, the ligands and downstream signaling of these receptors have remained a mystery. Two recent papers have now identified secreted R-spondin (RSPO) proteins as ligands for LGR5 and its homologues and have demonstrated that RSPO binding of LGR4/5/6 potentiates canonical Wnt-beta catenin signaling (1,2). LGR5 is a marker of stem cells in the base of intestinal crypts and in hair follicles and has been previously shown to be itself a canonical Wnt target gene in these cells. Moreover, significant data support LGR5+ stem cells as cells of origin for colorectal carcinoma and also implicate LGR5 as a mediator of tumor aggression. The combined data from the Liu and Clevers labs now suggest that by acting as an upstream potentiator of Wnt-beta catenin signaling, LGR5 promotes the proliferation and expansion of stem cell populations. Intriguingly, despite their close identity with FSH, LH and TSH, LGR5 and its homologues do not appear to function as GPCRs. The cumulative data from both groups indicate that RSPO-induced activation of LGR4/5/6 does not signal through G-proteins nor induce beta arrestin translocation. Rather, RSPO-binding of the leucine-rich N-terminal domains leads to an increase in the phosphorylation of the Wnt co-receptor LRP6, thereby upregulating activity of the Frizzled-LRP6 receptor complex and potentiating beta catenin activity. Although it is conceivable that other ligands might exist for LGR5 and its homologues, these recent reports indicate that RSPO-binding of LGR5 maintains stem cell proliferation through Wnt-beta catenin signaling in a manner that is independent of G-protein coupled signaling.
(1) Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. (2011)
R-spondins function as ligands of the orphan receptors Lgr4 and Lgr5 to regulate Wnt/{beta}-catenin signaling.
Proc Natl Acad Sci U S A. 108: 11452-7. [PMID: 21693646]
(2) de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, Stange DE, van Es J, Guardavaccaro D, Schasfoort RB, Mohri Y, Nishimori K, Mohammed S, Heck AJ, Clevers H. (2011)
Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling.
Nature. Published online ahead of print Jul 4 2011. DOI: 10.1038/nature10337. [PMID: 21727895]
IUPHAR review article published on the nomenclature, distribution and pathophysiological functions of Leukotriene receptors.
Bäck M, Dahlén S-E, Drazen JM, Evans JF, Serhan CN, Shimizu T, Yokomizo T, Rovati GE. (2011)
International Union of Basic and Clinical Pharmacology. LXXXIV: Leukotriene Receptor Nomenclature, Distribution, and Pathophysiological Functions.
Pharmacol Rev. 63:539-584 [Abstract]
IUPHAR review article published updating the classification of Prostanoid receptors.
Woodward DF, Jones RL, Narumiya S. (2011)
International Union of Basic and Clinical Pharmacology. LXXXIII: Classification of Prostanoid Receptors, Updating 15 Years of Progress.
Pharmacol Rev. 63:471-538 [PMID: 21752876]
IUPHAR review article published on the nomenclature and classification of Hydroxy-carboxylic Acid receptors (GPR81, GPR109A and GPR109B).
Offermanns S, Colletti SL, Lovenberg TW, Semple G, Wise A, IJzerman AP. (2011)
International Union of Basic and Clinical Pharmacology. LXXXII: Nomenclature and Classification of Hydroxy-carboxylic Acid Receptors (GPR81, GPR109A, and GPR109B).
Pharmacol Rev. 63: 269-90. [PMID: 21454438]
IUPHAR review article published on the nomenclature and classification of Adenosine receptors.
Fredholm BB, Ijzerman AP, Jacobson KA, Linden J, Müller CE. (2011)
International Union of Basic and Clinical Pharmacology. LXXXI. Nomenclature and Classification of Adenosine Receptors—An Update.
Pharmacol Rev. 63: 1-34. [PMID: 21303899]
Progesterone puts a swing in the tail.
Comments by David E. Clapham and John A. Peters:
Two recent reports in Nature by Strünker et al. (1) and Lishko et al. (2) have answered a long standing question in reproductive physiology: how does progesterone cause a rapid influx of Ca2+ into human spermatozoa? Using patch-clamp recording from human mature sperm cells (1,2) and optical techniques (1) the Authors provide compelling evidence that progesterone causes the activation and potentiation of a class of calcium selective ion channel that is activated by depolarization and which is expressed exclusively in the testes and sperm, namely the CatSpers (3). CatSpers are assembled as a complex of pore-forming CatSper1-4 subunits in association with CatSperβ, γ and δ auxiliary subunits, all of which are essential for function (4). Intracellular alkalinization of sperm, as occurs in the female reproductive tract, causes the opening of CatSper channels triggering Ca2+ entry and hyperactivation (whip-like flagellar beats) that are necessary for penetration of the egg cumulus and zona pellucida and subsequent fertilization (4). Alkalinization causes a hyperpolarizing shift in the voltage dependency of CatSper opening, an action that the recent reports (1, 2) also find for low nanomolar concentrations of progesterone, which acts in synergy with increased intracellular pH to stimulate CatSper mediated Ca2+ influx. Crucially, all of the evidence points to non-genomic action of progesterone via a cell surface receptor and, furthermore, to one that does not involve second messenger signalling.
The molecular target of progesterone remains uncertain: the possibilities include the CatSper complex itself, or an associated protein. These studies (1, 2) expand the list of ion channels that are subject to non-genomic regulation by steroid hormones. They also identify an interesting species difference in sperm regulation, since mouse CatSper activity is not increased by progesterone (2). Mechanistically, it is intriguing that the voltage-dependency of the opening of human and mouse CatSper differs substantially (2). Identifying the receptor for progesterone that modulates CatSper may potentially reveal a target for a novel male contraceptive agent in man.
(1) Strünker T, Goodwin N, Brenker C, Kashikar ND, Weyland I, Seifert R. (2011)
The CatSper channel mediates progesterone-induced Ca2+ influx in human sperm.
Nature. 471: 382-386. [PMID: 21412338]
(2) Lishko PV, Botchkina IL, Kirichok Y. (2011)
Progesterone activates the principal Ca2+ channel of human sperm.
Nature. 471: 387-391. [PMID: 21412339]
(3) Clapham DE, Garbers DL. (2005)
International Union of Pharmacology. L. Nomenclature and structure-function relationships of CatSper and two-pore channels.
Pharmacol Rev. 57: 451-454. [PMID: 16382101]
Three papers explore the structures of agonist-bound β-adrenoceptors.
(1) Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. (2011)
Structure and function of an irreversible agonist-β(2) adrenoceptor complex.
Nature. 469: 236-40 [PMID: 21228876]
(2) Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. (2011)
Structure of a nanobody-stabilized active state of the β(2) adrenoceptor.
Nature. 469: 175-80 [PMID: 21228869]
(3) Warne T, Moukhametzianov R, Baker JG, Nehmé R, Edwards PC, Leslie AG, Schertler GF, Tate CG. (2011)
The structural basis for agonist and partial agonist action on a β(1)-adrenergic receptor.
Nature. 469: 241-4 [PMID: 21228877]
Commentary in:
(4) Sprang SR. (2011)
Cell signalling: Binding the receptor at both ends.
Nature. 469: 172-3 [PMID: 21228868]
(5) Nature news article from 12th January 2011: "Near-action shots of vital proteins".
An update paper on IUPHAR-DB is published in the 2011 Nucleic Acids Research Database Issue.
Sharman JL, Mpamhanga CP, Spedding M, Germain P, Staels B, Dacquet C, Laudet V, Harmar AJ, and NC-IUPHAR. (2011)
IUPHAR-DB: new receptors and tools for easy searching and visualization of pharmacological data.
Nucl. Acids Res. 39 (Database Issue): D534-D538. [Abstract] [Full text]
2010
A population-specific HTR2B stop codon predisposes to severe impulsivity.
Comments by A.J. Harmar:
Bevilacqua and colleagues (1) identified a single nucleotide polymorphism in the gene encoding the 5-HT2B receptor (HTR2B Q20*) that was significantly associated with impulsivity in a Finnish population of violent offenders and matched controls. The polymorphism, which was only found in Finnish populations, introduces a stop codon into the N-terminal extracellular domain of the receptor, leading to reduced expression of the 5-HT2B receptor protein in the brain. 5-HT2B receptor knockout (Htr2b-/-) mice have reduced viability due to cardiovascular defects, but those that survive have a normal lifespan (2). These mice displayed increased impulsive behaviour, according to several measures.
(1) Bevilacqua L, Doly S, Kaprio J, Yuan Q, Tikkanen R, Paunio T, Zhou Z, Wedenoja J, Maroteaux L, Diaz S, Belmer A, Hodgkinson CA, Dell'osso L, Suvisaari J, Coccaro E, Rose RJ, Peltonen L, Virkkunen M, Goldman D. (2010)
A population-specific HTR2B stop codon predisposes to severe impulsivity.
Nature. 468: 1061-1066. [PMID: 21179162]
(2) Nebigil CG, Choi DS, Dierich A, Hickel P, Le Meur M, Messaddeq N, Launay JM, Maroteaux L. (2000)
Serotonin 2B receptor is required for heart development.
Proc Natl Acad Sci U S A. 97: 9508-9513 [PMID: 10944220]
Crystal structure of the human dopamine D3 receptor in complex with the small molecule D2/D3-specific antagonist eticlopride.
Chien EYT, Liu W, Zhao Q, Katritch V, Won Han G, Hanson MA, Shi L, Hauck Newman A, Javitch JA, Cherezov V, Stevens RC. (2010)
Structure of the Human Dopamine D3 Receptor in Complex with a D2/D3 Selective Antagonist.
Science. 330 (6007): 1091-1095; DOI: 10.1126/science.1197410. [Abstract] [Full text]
IUPHAR review article published on the nomenclature, distribution and function of the Kisspeptin receptor.
Kirby HR, Maguire JJ, Colledge WH, Davenport AP. (2010)
International Union of Basic and Clinical Pharmacology. LXXVII. Kisspeptin Receptor Nomenclature, Distribution, and Function.
Pharmacol Rev. 62 (4): 565-78. [PMID:21079036]
IUPHAR review article published on the nomenclature of Lysophospholipid receptors.
Chun J, Hla T, Lynch KR, Spiegel S, Moolenaar WH. (2010)
International Union of Basic and Clinical Pharmacology. LXXVIII. Lysophospholipid Receptor Nomenclature.
Pharmacol Rev. 62 (4): 579-87. [PMID:21079037]
IUPHAR review article published on the nomenclature and pharmacology of Cannabinoid receptors.
Pertwee RG, Howlett AC, Abood ME, Alexander SP, Di Marzo V, Elphick MR, Greasley PJ, Hansen HS, Kunos G, Mackie K, Mechoulam R, Ross RA. (2010)
International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid Receptors and Their Ligands: Beyond CB1 and CB2.
Pharmacol Rev. 62 (4): 588-631. [PMID:21079038]
IUPHAR review article published on the structure, signalling, accessory proteins, receptor dynamics and pharmacology of Frizzled class receptors.
Schulte G. (2010)
International Union of Basic and Clinical Pharmacology. LXXX. The Class Frizzled Receptors.
Pharmacol Rev. 62 (4): 632-67. [PMID:21079039]
Crystal structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. (2010)
Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists.
Science. Published ahead of print Oct 7, 2010; DOI: 10.1126/science.1194396. [PMID:20929726]
Time-resolved FRET between GPCR ligands reveals oligomers in native tissues.
Albizu L, Cottet M, Kralikova M, Stoev S, Seyer R, Brabet I, Roux T, Bazin H, Bourrier E, Lamarque L, Breton C, Rives ML, Newman A, Javitch J, Trinquet E, Manning M, Pin JP, Mouillac B, Durroux T. (2010)
Time-resolved FRET between GPCR ligands reveals oligomers in native tissues.
Nat Chem Biol. 6 (8): 587-94. [PMID:20622858]
IUPHAR review article published on the physiology, pharmacology and pathophysiological function of TRP channels.
Wu LJ, Sweet TB, Clapham DE. (2010)
International Union of Basic and Clinical Pharmacology. LXXVI. Current progress in the mammalian TRP ion channel family.
Pharmacol Rev. 62 (3): 381-404. [PMID:20716668]
Official IUPHAR nomenclature and review article published on the Apelin Receptor
Pitkin SL, Maguire JJ, Bonner TI, Davenport, AP. (2010)
International Union of Basic and Clinical Pharmacology. LXXIV. Apelin Receptor Nomenclature, Distribution, Pharmacology, and Function.
Pharmacol Rev. 62 (3): 331-42. [PMID:20605969]
Official IUPHAR nomenclature and review article published on Melatonin Receptors
Dubocovich ML, Delagrange P, Krause DN, Sugden D, Cardinali DP, Olcese J. (2010)
International Union of Basic and Clinical Pharmacology. LXXV. Nomenclature, Classification, and Pharmacology of G Protein-Coupled Melatonin Receptors.
Pharmacol Rev. 62 (3): 343-80. [PMID:20605968]
2009
X-ray structure, symmetry and mechanism of an AMPA-subtype Glutamate Receptor
Comments by J A. Peters, G.L. Collingridge, M. Spedding and R.W. Olsen:
Ligand-gated ion channels (LGICs) exist as pentameric (i.e., nicotinic ACh, 5-HT3, GABAA and glycine), tetrameric (i.e., ionotropic glutamate) and trimeric (i.e., P2X) complexes. Although an almost complete medium resolution (4Å) structure of the nicotinic ACh receptor of Torpedo has been available for several years (1), it was only recently that a 3.1Å resolution crystal structure of a zebrafish P2X receptor was reported by the laboratory of Eric Gouaux (2). The same laboratory has now revealed in Nature (3) an almost complete 3.6Å resolution crystal structure of a representative of the third structural class of LGIC, the rat homotetrameric GluA2 receptor, in the closed state. The study confirms previous structures of the amino terminal domain (ATD) and ligand binding domain (LBD) obtained in isolation that is in each case arranged as a pair of dimers. Agonist/competitive antagonist binding sites are located within and not between subunits; this differs from the pentameric LGICs which have ligand binding sites at subunit interfaces (1). Remarkably, the new GluA2 receptor study reveals that crossover occurs between the ATD and LBD, such that subunit domains within the dimeric pairs swap. In addition, this structure allows a first glance of the ion channel, around which the subunits no longer exist in pairwise arrangement, but become independent and adopt a four-fold symmetry. The regions of the polypeptide linking the ATD to the LBD, and the latter to the transmembrane domains, are also revealed for the first time in this study, and will no doubt prove important for analyzing mechanisms both of agonist-gated channel opening and desensitization, as well as modulation by allosteric ligands. The laboratory of Eric Gouaux had previously reported the structural basis of desensitization (4) and of partial agonism (5) at the same receptors, and these reports were already of great interest for drug design, in this competitive area. This report is certain to initiate a flurry of experimental activity.
(1) Unwin N. (2005).
Refined structure of the nicotinic acetylcholine receptor at 4Å resolution.
J Mol Biol. 346: 967-989. [PMID: 15701510]
(2) Kawate T, Michel JC, Birdsong WT, Gouaux E. (2009).
Crystal structure of the ATP-gated P2X4 ion channel in the closed state.
Nature. 460: 592-598.[PMID: 19641588]
(3) Sobolevsky AI, Rosconi MP, Gouaux E. (2009).
X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor.
Nature. 462: 745-756. [PMID: 19946266]
(4) Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. (2003).
Structural basis for partial agonist action at ionotropic glutamate receptors.
Nat Neurosci. 6: 803-10. [PMID: 12872125]
(5) Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. (2002).
Mechanism of glutamate receptor desensitization.
Nature. 417: 245-53.[PMID: 12015593]
α2A-adrenergic receptor contributes to Type 2 diabetes
Comments by R.R. Neubig:
Renström and colleagues report in Science Express that overexpression of the α2A-adrenergic receptor, which is encoded by a gene within a region of rat chromosome 1 (Niddm1) that influences susceptibility to diabetes, contributes to the reduced insulin secretion and impaired glucose tolerance in diabetic GK rats. The alpha2 adrenergic blocker yohimbine markedly improved insulin secretion and glucose handling in the diabetic rats. A similar effect was also shown in humans, where SNPs upstream of ADRA2A are associated with reduced glucose-stimulated plasma insulin levels and increased receptor mRNA in islets. This study suggests that in a subset of diabetics, alpha2 blockers that act selectively in periphery could represent a novel therapeutic approach.
(1) Rosengren AH, Jokubka R, Tojjar D, Granhall C, Hansson O, Li DQ, Nagaraj V, Reinbothe TM, Tuncel J, Eliasson L, Groop L, Rorsman P, Salehi A, Lyssenko V, Luthman H, Renström E. (2010)
Overexpression of Alpha2A-Adrenergic Receptors Contributes to Type 2 Diabetes.
Science. 327 (5962): 217-20. [PMID: 19965390]
Official IUPHAR nomenclature and review article published on formyl peptide receptors
Ye RD, Boulay F, Wang JM, Dahlgren C, Gerard C, Parmentier M, Serhan CN, Murphy PM. (2009)
International Union of Basic and Clinical Pharmacology. LXXIII. Nomenclature for the formyl peptide receptor (FPR) family.
Pharmacol Rev. 61 (2): 119-61. [PMID:19498085]
Official IUPHAR nomenclature and review article published on trace amine receptor
Maguire JJ, Parker WA, Foord SM, Bonner TI, Neubig RR, Davenport AP. (2009)
International Union of Pharmacology. LXXII. Recommendations for trace amine receptor nomenclature.
Pharmacol Rev. 61 (1): 1-8. [PMID:19325074]
2008
Official IUPHAR nomenclature and review article published on free fatty acid receptors
Stoddart LA, Smith NJ, Milligan G. (2008)
International Union of Pharmacology. LXXI. Free fatty acid receptors FFA1, -2, and -3: pharmacology and pathophysiological functions.
Pharmacol Rev. 60 (4): 405-17. [PMID:19047536]
Revised recommendations for nomenclature of ligand-gated ion channels
The nomenclature of ligand-gated ion channels and their subunits has recently been re-examined by NC-IUPHAR. Their revised recommendations for nomenclature are summarised here.
Crystal Structure of a human A2A Adenosine Receptor
Comments by S.P.H. Alexander, T.I. Bonner and A. Christopoulos:
Following on from reports of β-adrenoceptor structures reported recently, the 2.6 Å crystal structure of a further Gs-coupled receptor has been reported. The A2A receptor was modified, replacing the third intracellular loop with T4 bacteriophage lysozyme and deleting the C-terminus after the initial 25-30 residues beyond TM7. Purification in the presence of theophylline, which was later exchanged for the more selective A2A receptor antagonist ZM241385 allowed diffraction data to be obtained from the best 13 crystals. From the resulting solved structure, there were three main findings of particular note. The first is the presence of 4 disulfide bonds in the extracellular loop regions, which yields an organization that is very different from previously solved structures of rhodopsin and the β-adrenoceptor structures. Second, the transmembrane helices diverge from the orientations adopted by the corresponding domains in the rhodopsin and adrenoceptor structures. Finally, and perhaps most strikingly, these structural features result in a binding mode of the antagonist that places it in an extended conformation, almost perpendicular to the plane of the membrane, lined up against TM7 and interacting with the loop regions. This pose is very different to that predicted previously based on homology models.
(1) Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. (2008)
The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science. Nov 21; 322 (5905): 1211-7. [PMID: 18832607]
Structure of the β1-adrenergic receptor
Comments by A.J. Harmar:
Schertler and colleagues report the crystal structure of a β1-adrenergic receptor in complex with the antagonist cyanopindolol. Site directed mutagenesis was used to improve the thermostability of the protein and lock it in the antagonist state. This approach may be a fruitful one for determining the structures of other GPCRs.
(1) Warne T, Serrano-Vega MJ , Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tate CG, Schertler GFX. (2008)
Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. Jul 24; 454 (7203): 486-91 [PMID: 18594507]
2007
Crystal structure of human β2-adrenergic receptor
Comments by A.P.Davenport:
To date, only 148 unique structures for membrane proteins have been determined, only 4 of these are human in origin and only one crystal structure of a GPCR has been solved, the visual sensory protein rhodopsin. Three papers in Science and Nature now report the structure of the human β2-adrenergic receptor.
(1) Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. (2007)
Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. Nov 15; 450 (7168): 383-7.
[PMID: 17952055]
(2) Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ,
Kuhn P, Weis WI, Kobilka BK, Stevens RC. (2007)
High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. Nov 23; 318 (5854): 1258-65.
[PMID: 17962520]
(3) Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. (2007)
GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. Nov 23; 318 (5854): 1266-73. [PMID: 17962519]






